Abstract
Studies of hybridizing species can reveal much about the genetic basis and maintenance of species divergence in the face of gene flow. Here we report a genetic segregation and linkage analysis conducted on F2 progeny of a reciprocal cross between Senecio aethnensis and S. chrysanthemifolius that form a hybrid zone on Mount Etna, Sicily, aimed at determining the genetic basis of intrinsic hybrid barriers between them. Significant transmission ratio distortion (TRD) was detected at 34 (∼27%) of 127 marker loci located in nine distinct clusters across seven of the ten linkage groups detected, indicating genomic incompatibility between the species. TRD at these loci could not be attributed entirely to post-zygotic selective loss of F2 individuals that failed to germinate or flower (16.7%). At four loci tests indicated that pre-zygotic events, such as meiotic drive in F1 parents or gametophytic selection, contributed to TRD. Additional tests revealed that cytonuclear incompatibility contributed to TRD at five loci, Bateson–Dobzhansky–Muller (BDM) incompatibilities involving epistatic interactions between loci contributed to TRD at four loci, and underdominance (heterozygote disadvantage) was a possible cause of TRD at one locus. Major chromosomal rearrangements were probably not a cause of interspecific incompatibility at the scale that could be examined with current map marker density. Intrinsic genomic incompatibility between S. aethnensis and S. chrysanthemifolius revealed by TRD across multiple genomic regions in early-generation hybrids is likely to impact the genetic structure of the natural hybrid zone on Mount Etna by limiting introgression and promoting divergence across the genome.
Keywords: genetic divergence, genetic maps, genomic incompatibility, hybrid zone, intrinsic hybrid barrier, transmission ratio distortion
Introduction
Although strong, divergent natural selection can maintain population divergence in the face of gene flow (Nosil, 2012), intrinsic genetic barriers can also evolve under such conditions (Rundle and Nosil, 2005; Agrawal et al., 2011) or may already be in place to varying degrees between hybridizing populations that diverged in allopatry (Coyne and Orr, 2004; Bierne et al., 2011; Feder et al., 2012; Abbott et al., 2013). Intrinsic incompatibility between species is generally detected by crossing studies, which can also reveal the nature of such incompatibility at both genetic and genomic levels. For example, transmission ratio distortion (TRD), which is often observed at segregating loci among progeny of interspecific crosses or crosses between divergent lineages within species, can result from selection against particular hybrid genotype combinations at these loci (TRDLs; Fishman et al., 2001; Moyle and Graham, 2006). TRD may result from Bateson–Dobzhansky–Muller (BDM) incompatibilities at haploid and/or diploid stages of the life cycle caused by negative epistatic interactions between nuclear loci showing polymorphisms within and between populations of the same or different species (Fishman et al., 2008; Cutter, 2012; Bomblies, 2013; Corbett-Detig et al., 2013; Ouyang and Zhang, 2013). It may also arise from cytonuclear incompatibility (Levin, 2003; Fishman and Willis, 2006; Turelli and Moyle, 2007) or from ‘selfish' meiotic drive of alleles in a new genomic background (Fishman and Willis, 2005). Incompatibility caused by cytonuclear or haploid–diploid incompatibilities may depend on cross direction leading to asymmetric incompatibility, which can influence patterns of introgression following hybridization (Fishman et al., 2001; Turelli and Moyle, 2007; Tang et al., 2010). Genetic incompatibilities of all kinds accumulate with increasing phylogenetic distance until complete reproductive isolation is evident (Matute et al., 2010; Moyle and Nakazato, 2010; Levin, 2012; Corbett-Detig et al., 2013).
Interspecific crossing, when used in linkage analysis and mapping, may also reveal chromosomal rearrangements between species that can reduce the fitness of hybrids and suppress recombination that, in turn, will affect rates of interspecific gene flow (Rieseberg, 2001; Ortiz-Barrientos et al., 2002; Kirkpatrick and Barton, 2006). Studies of hybridizing annual sunflowers have revealed that differences in genetic architecture due to chromosomal inversions and translocations are an important cause of hybrid sterility (Lai et al., 2005; Yatabe et al., 2007), whereas in other groups of hybridizing species such as irises (Taylor et al., 2012) and sculpin fish (Stemshorn et al., 2011) such rearrangements appear to be absent or minor. In contrast, the presence of TRDLs in genetic maps of interspecific crosses appears to be common, if not the rule (Fishman et al., 2001; Lu et al., 2002; Tang et al., 2010). Either way, the cumulative action of multiple intrinsic incompatibilities with diverse modes of action distributed across the genome will act as a potent barrier to gene flow between species and seems to be a common, if not universal, intermediate stage in the process of speciation (Bierne et al., 2011; Feder et al., 2012; Abbott et al., 2013; Bomblies, 2013).
In the present study, we utilize interspecific crossing and linkage analysis to investigate the genetic nature of intrinsic incompatibility between two diploid, ragwort species, Senecio aethnensis (2n=20) and S. chrysanthemifolius (2n=20) (Asteraceae), that form a hybrid zone on Mount Etna, Sicily (James and Abbott, 2005; Brennan et al., 2009). These two species are self-incompatible, short-lived, herbaceous perennials, which grow at high and low altitudes, respectively, on Mount Etna. They are connected by a series of hybrid populations, which potentially provides a corridor for high levels of interspecific gene flow to occur. Interestingly, material collected from this hybrid zone and introduced to Britain in the late 17th century subsequently gave rise to a highly invasive homoploid hybrid species, S. squalidus, which spread through much of Britain in the 19th and 20th centuries (James and Abbott, 2005; Abbott et al., 2009).
Previous analyses of the hybrid zone on Mount Etna showed that while extrinsic environmental selection is important in determining the ecological differences and relative distributions of the two Senecio species (Brennan et al., 2009; Ross et al., 2012; Chapman et al., 2013; Muir et al., 2013; Osborne et al., 2013), intrinsic selection against hybrids was predominant in determining changes in allele frequencies and quantitative trait expression in the hybrid zone (Brennan et al., 2009). Indeed, based on indirect measures of assessment, Brennan et al. (2009) showed that the hybrid zone was characterized by strong selection against hybrids, high dispersal rates and few loci differentiating quantitative traits. The evidence for strong selection against hybrids was somewhat surprising, as both Hegarty et al. (2009) and Brennan et al. (2013) have reported that fertile hybrids are easily produced from crosses between the two species, whereas Chapman et al. (2005) showed that S. aethnensis exhibits no conspecific pollen advantage when pollinated with mixtures of pollen from both species and S. chrysanthemifolius showed only a small conspecific pollen advantage when treated similarly. However, Hegarty et al. (2009) noted that a marked decline in germination rate and survival occurred in the F2 generation of crosses they examined, indicating that post-mating incompatibility between the two species becomes apparent after the F1 generation, i.e., as a consequence of hybrid breakdown.
Clearly, the two Senecio species and the hybrid zone they form on Mount Etna comprise a very useful system for investigating adaptive divergence and mechanisms of reproductive isolation between hybridizing, diploid species. In the present study, we investigate further the nature of intrinsic reproductive isolation between S. aethnensis and S. chrysanthemifolius by examining genetic segregation in an F2 population derived from a reciprocal cross between the two species. This enabled us to (i) construct a genetic linkage map based on segregation in this F2 population, (ii) identify if large-scale linkage group (chromosomal) rearrangements exist between the two species, (iii) determine the occurrence and extent of TRD across the linkage groups identified and (iv) examine some of the possible causes of TRD at particular loci. Overall, the aim of our study was to obtain a better understanding of the occurrence and genetics of intrinsic incompatibility between these two species.
Materials and methods
F2 population
Parent plants were raised from seed collected from wild populations located at ∼2600 m (S. aethnensis population VB) and 600 m (S. chrysanthemifolius population C1) altitude, respectively, on Mount Etna, Sicily (see James and Abbott (2005) for further details of populations). Reciprocal crosses (Brennan et al., 2013) were made between one representative of each of S. aethnensis (A) and S. chrysanthemifolius (C) to produce 16 F1 individuals. The F1s were grown to flowering and inter-crossed in a partial diallel design. The most compatible reciprocal cross between a pair of F1s, i.e., producing highest seed-set in both directions, was chosen to found an F2 reciprocal cross family (hereafter referred to as F2AC). The maternal and paternal origin of each F2 individual and its F1 progenitors were recorded to permit testing of cytoplasmic effects. A total of 120 F2AC seed was sown, half coming from each direction of a cross between the selected pair of F1 parents. Following germination, F2AC individuals were grown, one per pot (15 cm diameter, containing a 3:1 compost to grit mix), in a glasshouse. Pots were randomized on a bench and re-randomized weekly until commencement of flowering. Twenty F2AC individuals (16.7%) failed to flower because they either failed to germinate, experienced early mortality or exhibited stunted growth and remained vegetative during the course of the study. As plants comprising this F2 family were also to be used in another study aimed at examining the quantitative genetics of morphological and life history differences between S. aethnensis and S. chrysanthemifolius, non-flowering individuals were excluded from further analysis. This left 100 F2 plants for genotyping and use in the construction of linkage maps.
DNA isolation and genotyping
DNA was extracted from young fresh leaves of all plants, i.e., the two parents, 16 F1 progeny, 100 F2AC progeny (Brennan et al., 2009). For a subsample of 9.4% of randomly chosen plants (41 out of 436 plants from a larger genetic study), two independent DNA extracts were made to test for genotyping reliability.
AFLP genotyping
A protocol modified from that described by Wolf (http://archive.is/LCe6) was used to generate amplified fragment length polymorphisms (AFLPs; Supplementary Methods). Fluorescently labelled forward primers were multiplexed in the final selective PCR step by adding 0.05 μl of each of two labelled primers per sample and adjusting the total 20 μl PCR volume accordingly.
The AFLP protocol was initially tested on a panel of individuals comprising the two parents and their F1 progeny. AFLPs were detected by running samples on a Beckman Coulter CEQ 8000 capillary sequencer (Brennan et al., 2009) and analysing output with CEQ v9.0 (Beckman Coulter Inc., Fullerton, CA, USA, 2004). In total, 28 paired combinations of five selective forward and eight selective reverse AFLP primers with different combinations of three final 5′ bases were tested on these individuals. Samples were genotyped first using the automatic AFLP binning options of CEQ v9.0 with a minimum relative fluorescence unit (r.f.u.) cut-off of 100 r.f.u. and bin widths of 1 bp. Manual checks of the AFLP genotypes of parents, repeat extracts, and F1 plants were then conducted to assess what subset of AFLP bands were suitable for genotyping F2AC individuals based on reliable amplification and inheritance pattern. Eight of the 28 primer combinations (E1M3, E1M5, E1M7, E4M7, E5M3, E5M6, E8M5, E8M7) were chosen for genotyping F2AC plants because they produced many reliably scored amplified polymorphic bands showing high levels of repeatability across replicate extracts (>95% across all scored loci) and the expected inheritance pattern from parents to F1s, and also because they could be used in multiplexed combinations (Supplementary Table S2a). AFLP bands were scored in F2AC individuals from the automatic AFLP CEQ v9.0 binning output with reference to parent controls and then checked manually across all samples to correct for genotyping errors.
SSR, EST and INDEL genotypes
A screen of 340 existing and newly developed single-locus molecular genetic markers was initially carried out on the two parent individuals. These loci comprised 37 simple sequence repeat loci (SSRs) developed previously by repeat motif enrichment of genomic DNA of S. aethnensis, S. chrysanthemifolius, S. squalidus and S. vulgaris (Liu et al., 2004), 25 newly developed SSRs (A Brennan and G-Q Liu, unpublished), 10 SSRs developed for S. madagascariensis (Le Roux and Wieczorek, 2006), 216 newly developed expressed sequence tag (EST)-SSRs and 45 EST-indels (SSR markers labelled as ES and NES, indels labelled as EC; Hegarty et al., 2008; www.seneciodb.org/), and 7 additional indel markers comprising five derived from non-duplicated Asteraceae genes (Chapman et al., 2007) and two from published polymorphic Senecio gene sequences (SSP, which encodes a stigma-specific peroxidase involved in pollination (McInnis et al., 2005), and Ray2a, which encodes a cycloidea-like transcription factor involved in the control of ray floret development in Senecio (Kim et al., 2008)). A single PCR protocol was used to amplify all markers (Brennan et al., 2009). This PCR protocol consisted of a three-primer system with universal fluorescently labelled M13 primers. Mulitplexed PCR products of all F2 plants, wild sampled plants, control repeats, and both parents were run on a Beckman Coulter CEQ 8000 capillary sequencer and genotypes were assessed and scored using CEQ v9.0. Fifty-four of these marker loci were appropriate for genetic mapping in the current study based on reliable PCR amplification, fragment length scoring and the presence of fragment length polymorphisms in the F2AC family (Table 1 and Supplementary Table S1).
Table 1. Summary of genetic markers screened and mapped in the Senecio aethnensis × S. chrysanthemifolius F2 genetic mapping family.
| Marker type | No. of screened | No. of developed | No. of genotyped | No. of mapped | Unlinked | Problematic |
|---|---|---|---|---|---|---|
| Codominant markers | ||||||
| EST indel | 45 | 15 | 10 | 8 | EC77, EC1687 | |
| EST SSR | 216 | 48 | 33 | 31 | ES91 | ES19 |
| Indel | 7 | 4 | 3 | 3 | ||
| SSR | 72 | 9 | 8 | 8 | ||
| Total | 340 | 76 | 54 | 50 | 1 | 3 |
| Dominant markers | ||||||
| E1M3, CAAC/ACAG | — | — | 19 | 15 | 179, 219 | |
| E1M5, CAAC/ACTA | — | — | 15 | 10 | 168, 213 | 88, 160, 204 |
| E1M7, CAAC/ACTG | — | — | 13 | 12 | 275 | |
| E4M7, CACT/ACTG | — | — | 9 | 8 | 113 | |
| E5M3, CACC/ACAG | — | — | 8 | 7 | 302 | |
| E5M6, CACC/ACTC | — | — | 9 | 9 | ||
| E8M5, CAGG/ACTA | — | — | 11 | 9 | 114 | 196 |
| E8M7, CAGG/ACTG | — | — | 7 | 7 | ||
| Total | — | — | 91 | 77 | 8 | 6 |
Abbreviations: EST, expressed sequence tag; SSR, simple sequence repeat.
Marker types are divided into codominant or dominant markers. Codominant markers are further divided into markers derived from ESTs, SSRs or insertion-deletions (indel). Dominant markers are amplified fragment length polymorphisms (AFLPs). Each AFLP primer combination is shown as the primer names used in this study followed by the three selective bases for the EcoRI and MseI primers, respectively (Supplementary Methods). No. of screened is number of codominant marker primer pairs tested. No. of developed is number of codominant markers that could be scored in Senecio. No. of genotyped is number of markers showing polymorphism in the F2AC mapping family. No. of mapped is number of markers that were included in the final genetic map. Unlinked names markers that were unlinked at a >4 logarithm of odds score or>20 cM linkage threshold limits for mapping. Problematic refers to markers that caused problems with linkage group marker order or were present in multiple linkage groups. Dominant marker names are approximate fragment base-pair lengths used to label AFLP loci.
Genetic mapping
A genetic linkage map was constructed from the segregation of alleles at marker loci in the F2AC family using the demonstration version of Joinmap v4.0 (Van Ooijen, 2001). In the analysis, the F2AC was treated as an outcrossed mapping family (CP type) because many AFLP and codominant marker genotypes were heterozygous in the F0 parents (S. aethenensis and S. chrysanthemifolius) and therefore did not exhibit the type of F2 inheritance that assumes parents are homozygous for alternative alleles at polymorphic loci. The outcross mapping family option (CP type) in Joinmap v4.0 allows mapping of loci with a variety of parental genotypes showing different segregation patterns. The software automatically assigns the most likely linkage phase of heterozygous parental alleles to each of the two parental chromosomes for each group of linked loci, thus enabling more loci to be mapped. A genetic map was assembled using Joinmap's default regression mapping algorithm parameters. Linkage groups were identified at greater than four logarithm of odds (LOD) score with less than 20 Kosambi centiMorgan (cM) map distance units between loci. Map quality was assessed by examining goodness of fit G2 statistics and markers responsible for incompatible linkage interactions were removed to generate linkage groups with high map support. In some cases, removal of suspect loci led to the splitting of large linkage groups. Diagrams of linkage groups were constructed using MapChart v2.2 (Voorrips, 2002).
Summary statistics describing map characteristics were calculated as follows. Genome length was estimated by adding twice the mean marker distance to the length of each linkage group to account for ends beyond the terminal markers (Fishman et al., 2001) and also by multiplying the length of each linkage group by the correction factor (marker number +1)/(marker number −1) (Chakravarti et al., 1991). Map coverage in terms of the percentage of the genome that is within 5 or 10 cM of a mapped marker was assessed according to the formula: 1−exponent((−2 × distance × marker number)/map length)) (Fishman et al., 2001). The extent of marker clustering was tested using a chi-square dispersion test against a null Poisson distribution of evenly distributed markers separated by mean marker distance.
Transmission ratio distortion
Genotype frequencies at each mapped marker locus in the entire F2AC mapping family were tested for Mendelian segregation of genotypes using chi-square tests with Microsoft Excel 2003 (Microsoft Corp). Associations between chi-square statistics for genotype segregation and marker characteristics, including marker type (classified as dominant or codominant) and AFLP fragment length, were tested with linear models after natural log transformation using R v2.13 (R Development Core Team, 2011).
The distribution of loci showing TRD, suggestive of the number of independent TRDLs, was examined by plotting per locus chi-square values onto their genetic map positions. The number of distinct TRDLs across the genetic map was assessed as the number of clusters of distorted markers at a single-locus 95% confidence level. Clustered distorted markers within each TRDL were located at a map distance of less than 10 cM from each other. The most likely map position of a TRDL was interpreted as the map position of the locus exhibiting the greatest TRD within each cluster of distorted markers. Some markers showing TRD were isolated by more than 10 cM from the nearest marker also exhibiting TRD. These markers were considered as possibly representing different TRDLs, although with weaker supporting evidence. Clustering of distorted markers across the entire genetic map was tested with a binomial test of the hypothesis that the observed number of neighbouring pairs of distorted loci per chromosome was greater than the expected number of pairs of such loci given their observed frequency. A Poisson test of TRDL clustering was not conducted, as the underlying marker distribution was significantly clustered according to a dispersion test. A possible bias of TRDLs being located at the ends of linkage groups, due possibly to weak linkage to all other mapped markers, was investigated using a binomial test of whether loci at linkage group ends more often exhibited segregation distortion relative to the overall likelihood of loci showing TRD.
Causes of TRD
Post-zygotic mortality or failure to flower
Chi-square tests were performed on the F2AC family of segregating offspring to test the extent to which selective post-zygotic mortality or failure to flower (exhibited by 16.7% of F2 individuals examined) could be responsible for generating the observed TRD. For these tests, expected genotype and allele segregation patterns were based on the minimum frequencies expected assuming that all non-genotyped F2 individuals possessed the under-represented allele or genotype.
Pre-zygotic events
TRD caused by pre-zygotic events, such as biased gamete production (meiotic drive), gametophyte selection or other maternal effects (Turelli and Moyle, 2007; Fishman et al., 2008), was tested at 36 codominant loci where the parental origin of alleles could be identified unambiguously. These early reproductive events occur at the haploid stage of the life cycle and require tests on allelic rather than genotype segregation. Because such modes of incompatibility may show unilateral effects and therefore be visible in only one cross direction, allelic segregation tests were also performed on subsets of F2 individuals containing different parental cytoplasms.
Cytonuclear incompatibility
Because the F2AC family was generated from a reciprocal cross, it was possible to examine whether TRD of genotypes at particular loci resulted from cytonuclear incompatibility by testing if TRD was dependent on the direction of the cross (Fishman and Willis, 2006; Turelli and Moyle, 2007). Thus, we tested TRD of genotypes in separate subsets of 49 and 51 individuals in the F2AC mapping family that had inherited either the S. aethnensis or S. chrysanthemifolius cytoplasm, respectively (Supplementary Figures S2 and S4).
BDM incompatibilities
Hybrid incompatibilities are often thought to be due to epistatic BDM incompatibilities between parental alleles at two or more interacting genetic loci. To test for this, interactions between all pairs of identified TRDLs were examined using Fisher's exact tests of contingency tables of paired genotype counts using R v2.13 (R Development Core Team 2011).
Deficiency or excess of heterozygotes
Tests of whether TRD was caused by either a deficiency or excess of heterozygous genotypes were conducted at 36 codominant loci where the heterozygous state of progeny in terms of parental alleles could be identified. A deficiency of heterozygous genotypes might reflect negative allelic interactions at a locus (underdominance). In contrast, an excess of heterozygous genotypes could reflect inbreeding depression (due to the expression of deleterious recessives) as a consequence of the full-sib cross used to generate the F2 family (Remington and O'Malley, 2000; Schwarz-Sommer et al., 2003), heterosis exhibited by heterozygous genotypes generated from parents homozygous for different alleles (Latta et al., 2007), or the occurrence of recessive BDM incompatibilities across two or more loci (Fishman et al., 2008).
Locally reduced recombination
Genomic regions exhibiting limited local recombination might show enhanced associations with TRD because of the effects of chromosomal rearrangements on chromosomal segregation during meiosis, or because they are more likely by chance to show linkage to nearby loci under selection in hybrids (Fishman et al., 2013). We investigated this by testing for significant associations between local recombination rate and TRD using logistic regression of markers scored for various categories of TRD against marker distance. Different TRD categories were chosen to reflect different stringencies in defining TRDLs and TRDL map locations. These included categories where markers exhibited TRD detected by chi-square tests at (i) P=0.05 or (ii) P=0.001, (iii) where markers exhibited greatest TRD within each of the nine identified TRDLs and (iv) where markers showed greatest TRD within each of the four multi-locus TRDLs.
Results
Molecular genetic markers
Details of the AFLP, and codominant markers selected to genotype S. aethnensis and S. chrysanthemifolius parents, and their F1 and F2 offspring for construction of linkage maps are presented in Table 1 and Supplementary Table S1.
Genetic linkage map
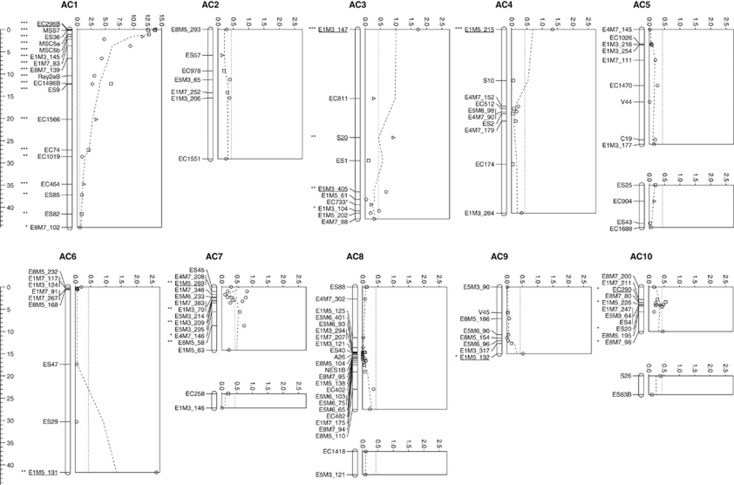
A genetic linkage map constructed from the segregation of markers in the F2AC mapping family comprised 127 marker loci distributed across 14 linkage groups of average 22.4 cM length (s.d., 15.6 cM) giving a total genetic map length of 313.8 cM (Figure 1 and Table 2). Markers showing TRD were included in the construction of this map because such distortion has been shown not to bias recombination statistics nor the resulting linkage maps (Xu, 2008). Nine markers were not mapped because they were unlinked at the <4 LOD and >20 cM thresholds for inclusion in linkage groups (Table 1). Another nine markers were excluded from the map because they caused problematic linkage interactions within linkage groups (Table 1) or, in the case of two of these markers, showed linkage to multiple different linkage groups (EC77 linked AC1 and AC5A, and EC1687 linked AC5A, AC5B, and AC6). Weak linkage (<4 LOD or >20 cM distances) between markers located on four pairs of linkage groups was attributed to them belonging to distant ends of the same chromosome. Thus, ten independent linkage groups were inferred, which matches the haploid chromosome number (n=10) of both species (Alexander, 1979). Distance between mapped markers averaged 2.8 cM, but was highly variable (s.d.=3.6 cM). The dispersion index for markers was large (4.66) and highly significant (dispersion test, P=5.85e-46), which indicated a clumped distribution of markers across the map. The total predicted map length was estimated to be 391.6 cM (Fishman et al., 2001) or 407.1 cM (using the method of Chakravarti et al. (1991)) and ∼96% and >99% map coverage of the genome was predicted to be within 5 and 10 cM of a mapped marker, respectively.
Figure 1.
Genetic map of a reciprocal F2 S. aethnensis and S. chrysanthemifolius mapping family showing mapped marker positions and associated TRD. Map distances in Kosambi centiMorgans are shown in the scale to the left of linkage groups. Linkage groups are represented by vertical bars with mapped locus positions indicated with horizontal lines. Weakly linked linkage groups (<4 LOD or >20 cM) that are thought to belong to the same chromosome are aligned vertically. Locus names are listed to the left of linkage groups. Asterisks to the left of locus names indicate significant transmission ratio distortion of genotype frequencies at a single locus χ2 test 5% confidence limit. Underlined locus names indicate a most strongly distorted locus within a particular cluster of distorted loci. Plots to the right of each linkage group show −log1000 P values for single locus χ2 tests for transmission ratio distortion of genotype frequencies. Plot symbols indicate the number of degrees of freedom for the χ2 tests; circles=1 df, squares=2 df, and triangles=3 df. The dotted lines indicate the moving average of three neighbouring loci while dashed lines indicate the 5% significance level, to the right of which, loci show significant transmission ratio distortion. The plot scale for linkage group AC1 is larger than the other linkage groups.
Table 2. Summary genetic linkage map statistics for the S. aethnensis × S. chrysanthemifolius F2 genetic map.
| Linkage group | Length | Total marker no. | Dominant marker no. | Codominant marker no. | Add2s length | Method4 length |
|---|---|---|---|---|---|---|
| 1 | 44.5 | 18 | 4 | 14 | 50.05 | 49.74 |
| 2 | 29.1 | 7 | 4 | 3 | 34.65 | 38.80 |
| 3 | 42.6 | 10 | 6 | 4 | 48.15 | 52.07 |
| 4 | 41.3 | 10 | 6 | 4 | 46.85 | 50.48 |
| 5A | 25.8 | 9 | 5 | 4 | 31.35 | 32.25 |
| 5B | 9.5 | 4 | 0 | 4 | 15.05 | 15.83 |
| 6 | 41.7 | 9 | 7 | 2 | 47.25 | 52.13 |
| 7A | 14.2 | 15 | 14 | 1 | 19.75 | 16.57 |
| 7B | 3.2 | 2 | 1 | 1 | 8.75 | 9.60 |
| 8A | 27.5 | 22 | 16 | 6 | 33.05 | 30.12 |
| 8B | 5.2 | 2 | 1 | 1 | 10.75 | 15.60 |
| 9 | 15 | 8 | 7 | 1 | 20.55 | 19.29 |
| 10A | 10 | 11 | 8 | 3 | 15.55 | 12.00 |
| 10B | 4.2 | 2 | 0 | 2 | 9.75 | 12.60 |
| Total | 313.8 | 127 | 77 | 50 | 391.56 | 407.06 |
| Mean | 22.41 | 9.07 | 5.50 | 3.57 | 27.97 | 29.08 |
| s.d. | 15.56 | 5.88 | 4.55 | 3.37 | 15.56 | 16.65 |
Map distance measures are in Kosambi centiMorgan units. Add2s length is an estimate of chromosome length calculated as linkage group length plus twice mean linkage group distance. Method4 length is another estimate of chromosome length calculated as linkage group length times (marker number+1)/(marker number −1).
Transmission ratio distortion
TRD was common for markers included in the F2AC genetic map with 34.0% of codominant markers and 22.1% of dominant markers showing significant deviations from Mendelian expectations of genotype segregation according to a per-locus 95% confidence level (26.8% overall percentage frequency; Figure 1 and Supplementary Table S2). Mapped codominant markers exhibited TRD more frequently than mapped dominant markers (F1,125=10.41, P=0.0016), probably reflecting the fact that AFLPs, but not codominant markers, were included on the basis of inheritance patterns in the F1 progeny panel. No association was found between AFLP fragment length and TRD (F1,75=0.67, P=0.4173).
The map locations of markers were investigated to better understand the genetic architecture of incompatibilities between the two parent species, S. aethnensis and S. chrysanthemifolius. Markers showing genotypic segregation distortion were clustered in the genetic map (one-way binomial tests P=0.0007 and 3.5e-6, markers showing TRD at individual chi-square test 95% and 99.9% confidence levels, respectively) to form four distinct groups located in linkage groups AC1, AC3 (distal position), AC7A, and AC10A, and also occurred individually in linkage groups AC3 (proximal and central widely separated positions), AC4, AC6 and AC9 (Figure 1 and Table 3). The widespread distribution of clusters and individual markers showing TRD across several linkage groups indicates that multiple loci contributed to the frequent TRD observed in the F2 of this cross (Figure 1, Supplementary Figure S1, Table 3 and Supplementary Table S2). Within each cluster, all markers showed the same TRD direction favouring either alleles from the same parent or heterozygous or homozygous parental genotypes (Supplementary Figures S1–S4). Markers located at the ends of six linkage groups exhibited significant TRD, which raised the possibility that such markers that are weakly linked to other mapped markers might be erroneously interpreted as distinct TRDLs. However, a binomial test for overrepresentation of linkage group ends exhibiting TRD (6 out of 28) relative to the overall frequency of markers showing segregation distortion (26.4%) was not significant (P=0.788). We conclude that at least four TRDLs influencing multiple markers were present in the mapping family or as many as nine TRDLs if additional unlinked markers showing TRD were also included.
Table 3. Summary of transmission ratio distortion loci and tests of different transmission ratio distortion mechanisms for the S. aethnensis × S. chrysanthemifolius F2 genetic mapping family.
| Linkage group | Reference marker | Map position (cM) | Late-acting sufficient | Asymmetric | Pre-zygotic | Heterozygote | Epistasis (minority genotype) |
|---|---|---|---|---|---|---|---|
| AC1 cluster | EC296 | 0.0 | No | No | aeth | Yes | No |
| AC3 singleton (proximal) | E1M3_147 | 0.0 | Yes | No | — | — | AC3 central (CA), AC6 (CD) |
| AC3 singleton (central) | S20 | 24.3 | No | chrys | chrys | No | AC3 proximal (AC), AC3 distal (AC), AC6 (AC) |
| AC3 cluster (distal) | E5M3_405 | 36.5 | Yes | chrys | — | — | AC3 central (CA), AC6 (DC) |
| AC4 singleton | E1M5_215 | 0.0 | No | No | — | — | No |
| AC6 singleton | E1M5_131 | 41.7 | No | No | — | — | AC3 proximal (DC), AC3 central (CA), AC3 distal (CD) |
| AC7 cluster (central) | E1M5_269 | 12.5 | No | No | — | — | No |
| AC9 singleton | E1M5_132 | 15 | No | chrys | — | — | No |
| AC10A cluster | EC290 | 3.4 | No | aeth | aeth | No | No |
| AC5Ba singleton | ES25 | 0.0 | — | aeth | — | — | — |
| AC10Ba singleton | S26 | 0.0 | — | No | aeth | No | — |
Abbreviations: TRD, transmission ratio distortion; TRDL, transmission ratio distortion loci.
Linkage group names correspond to Figure 1. Cluster or singleton refers to whether a cluster or a single marker within 10 cM was observed to exhibit genotypic TRD.
The ‘reference marker' is the marker with strongest TRD where a cluster of distorted markers was observed. ‘Late-acting sufficient' tests if selective removal of the minority genotype according to observed post-mortality and reproductive failure would be sufficient to explain observed TRDLs. ‘Asymmetric' tests if subsets of the mapping family divided according to parental cytotype showed asymmetric TRD in one cross direction only with ‘aeth' and ‘chrys' indicates if asymmetric TRD was observed in an S. aethensis or S. chrysanthemifolius cytoplasmic background, respectively. ‘Pre-zygotic' tests for allelic TRD with ‘aeth' and ‘chrys' indicating the parental alleles showing significant deficiencies and ‘—' indicating that no suitable codominant markers were available for testing at this TRDL. ‘Heterozygote' tests for significant excesses or defiencies in heterozygosity of genotypes scored according to parental allelic state with ‘—' indicating that no suitable codominant markers were available for testing at this TRDL. ‘Epistasis' tests for non-independence of paired TRDL reference marker genotypes, with significantly interacting TRDLs listed. Letter codes in parentheses indicate minority genotype for the reference TRDL followed by the interacting TRDL. Genotype codes: A, homozygous S. aethnensis alleles; C, not homozygous for S. aethnensis alleles; D, not homozygous for S. chrysanthemifolius alleles.
Indicates that the last two TRDLs were only observed when specific mechanisms were investigated, so are not presented in Figure 1 or counted as primary TRLDs in the text.
Possible causes of TRD
Post-zygotic mortality or failure to flower
The observed TRD could have been due to selective mortality or selective failure to flower of some individuals in the mapping family (16.7% of F2 progeny). Tests for TRD greater than could be accounted for by these causes showed that seven of the nine TRDLs (excluding the proximal and distal TRDLs on AC3) required additional mechanisms to explain the observed TRD in these regions (Table 3, Supplementary Table S2 and Supplementary Figure S1).
Pre-zygotic events
Tests of TRD caused by events at early stages of reproduction, such as meiotic drive and gametophytic selection, were conducted on codominant loci where the parental origin of alleles could be identified. These tests showed that for TRDLs in linkage groups AC1 and AC10A, and also for a new TRDL in AC10B, F2 progeny lacked S. aethnensis alleles, whereas for the central TRDL in linkage group AC3, F2 individuals lacked S. chrysanthemifolius alleles (Table 3, Supplementary Table S2 and Supplementary Figure S3). When cross direction was also tested for, TRDLs in AC5B and AC10A were shown to express TRD in the S. aethnensis maternal background only (Supplementary Figure S4 and Supplementary Table S2).
Cytonuclear incompatibility
Tests of TRD of genotypes in subsets of F2 individuals that reflected differences in cross direction showed an overall similar genomic distribution of TRD irrespective of parental cytoplasm (Supplementary Figures S2 and S4). However, the TRD of several TRDLs was cytoplasm dependent. This was the case for TRDLs located in linkage groups AC3 (central and distal positions) and AC9 (all dependent on S. chrysanthemifolius cytoplasm), and also the TRDL located in AC10A (dependent on S. aethnensis cytoplasm) (Table 3, Supplementary Table S2 and Supplementary Figure S2). A further instance of TRD in linkage group AC5B was detected only in progeny possessing S. aethnensis cytoplasm (Table 3 and Supplementary Figure S2). Taken overall, these tests found asymmetric TRD occurring at five TRDLs dependent on individuals possessing either S. chrysanthemifolius or S. aethnensis cytoplasm.
Epistatic BDM incompatibilities
Multi-locus BDM incompatibilities were investigated by looking for negative interactions between particular genotype combinations of pairs of markers close to the nine TRDLs identified. Of the 36 paired TRDL combinations tested, five significantly non-independent pairs were found that affected four of the nine TRDLs (all three TRDLs on AC3 and one on AC6; Table 3). However, two of the five interacting TRDLs were present in the same linkage group indicating that physical proximity might contribute to their non-independence. The under-represented genotype combinations typically included one of the loci homozygous for S. aethnensis alleles and the other locus homozygous for S. chrysanthemifolius alleles.
Deficiency or excess of heterozygotes
Tests of a deficiency or excess of heterozygotes in terms of parental alleles at codominant loci where the parental state of alleles could be identified, detected a significant deficiency of heterozygous genotypes at the TRDL of large effect located in linkage group AC1 (Table 3, Supplementary Table S2 and Supplementary Figure S1). However, this deficiency is more likely caused by selection against S. aethnensis alleles, than underdominance (heterozygote disadvantage), because there was an even stronger bias against genotypes homozygous for S. aethnensis alleles across this particular region of AC1 (Supplementary Figure S3). No TRDLs showing an excess of heterozygotes relative to homozygotes were identified, thus discounting inbreeding depression, heterosis, or recessive BDM incompatibilities, as important causes of TRD in this F2 family.
Locally reduced recombination
In no instance was an association detected between locally reduced recombination and TRD for any of the four categories of markers tested. Thus, for markers that showed TRD at 95% and 99.9% confidence levels, the probability of a stronger association with recombination rate than the null hypothesis of no association was P=0.300, and P=0.291, respectively, whereas the probability was P=0.261 for the most strongly distorted marker within each of the nine identified TRDLs, and P=0.178 for the most strongly distorted marker within each of the four multi-locus TRDLs treated separately. Thus, these results do not support the hypothesis that chromosomal regions with reduced recombination are associated with hybrid incompatibility in this system.
Discussion
Studies of closely related species that form hybrid zones can reveal much about the genetic basis and maintenance of species divergence in the face of frequent interspecific hybridization and gene flow. Our previous analysis of the hybrid zone between S. aethnensis (occurring at high altitude) and S. chrysanthemifolius (occurring at low altitude) on Mount Etna, Sicily, compared clines for molecular variation with those for phenotypic trait variation, and indicated that both extrinsic and intrinsic selection against hybrids act to maintain the hybrid zone despite high levels of gene flow (Brennan et al., 2009). The study reported here has expanded on these previous results and provided insights into the genetics of intrinsic reproductive isolation between S. aethnensis and S. chrysanthemifolius on Mount Etna, Sicily. We found intrinsic genomic incompatibility between these two species caused by a variety of genetic mechanisms at multiple genetic loci. However, large-scale genomic rearrangements or translocations between the species did not appear to contribute greatly to this incompatibility.
F2 linkage map structure
To investigate the genomic architecture of these hybridizing Senecio species, we constructed a genetic linkage map from the segregation of dominant and codominant molecular markers in the F2 of a reciprocal cross between the two species. The resulting F2AC linkage map comprised ten distinct linkage groups (taking account of four weakly linked linkage group pairs), which corresponds to the haploid chromosome number of the two species (Alexander, 1979). If the parental species were distinguished by chromosomal translocations, the affected regions would link different linkage groups in genetic maps of hybrids and reduce the number of independent linkage groups to below the haploid chromosome number of the species investigated (Fishman et al., 2013). Therefore, the ability to distinguish ten distinct linkage groups in the F2AC map and the removal from maps of only two markers with suspect linkage to multiple different linkage groups indicates that large-scale genomic translocations between chromosomes probably do not distinguish the two species in contrast to what has been found in hybridizing annual sunflowers (Lai et al., 2005; Yatabe et al., 2007).
Overall, the map showed good coverage with an average locus distance of just 2.8 cM and >99% of the genome predicted to be within 10 cM of a mapped marker. However, markers exhibited a highly clumped distribution within the map, for which there are several possible explanations. First, there may be technical reasons for marker clustering, such as marker position uncertainty due to a limited number of recombination events observed in the relatively small F2 family examined, and/or to genotyping error, which though estimated to be reasonably low (2.2% based on duplicated samples tested) translates to a 2.2 cM uncertainty in the position of loci within maps. Second, some marker clusters could be explained by sequence heterogeneity across the genome. For example, clusters of AFLP loci could signal repetitive genomic regions containing many closely spaced repeated restriction enzyme cut sites, whereas clusters of EST and gene loci could signal highly expressed, gene-rich genomic regions. Third, the occurrence of clusters could reflect variable recombination rates across the genome caused by intrinsic features such as low recombination near centromeres or in regions where local chromosome rearrangements, such as inversions, exist between species.
Transmission ratio distortion
A notable feature to emerge from the current study was the large number of molecular marker loci that showed TRD in the F2AC mapping population, i.e., 34 (26.8%) of 127 markers tested. These loci were non-randomly distributed across the genetic map and generally formed clusters in which all loci showed the same bias against alleles from one parent or against heterozygous combinations of parental alleles. We found strong multi-locus evidence for the occurrence of four TRDLs and weaker single-locus evidence for five additional TRDLs located in seven of the ten linkage groups identified. Individual markers showing TRD were sometimes located at the ends of linkage groups raising the possibility that they were technical artifacts; however, such markers were not over-represented relative to the overall observed numbers of markers exhibiting TRD.
One TRDL, located at the proximal end of linkage group AC1, showed particularly strong segregation distortion, both in terms of the extent to which allele and genotypes frequencies were distorted away from Mendelian expectations for S. aethnensis alleles and genotypes, and the length of the genome affected (all of AC1, that is, 44.5 cM). The fact that this TRDL, and also other multi-locus TRDLs, affected large genomic regions was likely due to both their strong effects on segregation distortion and the limited post-hybridization recombination that had occurred in the F2AC mapping family. Strong TRD in early-generation hybrids is likely to bias patterns of introgression across large genomic regions in later generation hybrids because hybrid genotypes at linked loci are eliminated before they have an opportunity to recombine away from the TRDL (Bierne et al., 2011).
Causes of TRD
Through further analyses of the patterns of segregation in the F2AC family, we obtained some insights into the causes of TRD at different TRDLs. Twenty of the original 120 progeny that comprised the F2AC mapping family failed to flower either because of a failure to germinate, the occurrence of early mortality, or an inability to develop to the flowering stage. These individuals were not genotyped and their absence may have contributed to the TRD observed at marker loci. The low intrinsic fitness of these individuals might stem from several causes (see below), including the disruption of coadapted gene complexes (hybrid breakdown) following recombination. We did not examine survival and flowering in the F1 generation in our study, however a previous study (Hegarty et al., 2009) recorded a marked drop in intrinsic fitness (measured in terms of seed germination and seedling survival under glasshouse conditions) between the F1 and F3 generations of a reciprocal cross between the same S. aethnensis and S. chrysanthemifolius, parental individuals as this study indicating the occurrence of hybrid breakdown.
Although post-zygotic selective mortality or inability to flower of some F2 individuals would contribute to TRD at certain marker loci, we found that the level of TRD exhibited by seven of the nine TRDLs identified could not be entirely explained in this way. However, it remains possible that earlier-acting post-zygotic incompatibility in the form of a failure of seed development (e.g. due to negative interactions between zygote and endosperm) could have contributed to the observed TRD. Alternatively, there may be pre-zygotic causes of TRD either in the production of gametes containing particular alleles (meiotic drive) or selection against gametes containing particular alleles (gametophytic selection) (Fishman and Willis, 2005; Fishman et al., 2008). We tested for these possibilities at loci where the parental origin of alleles could be identified unambiguously allowing TRD of alleles rather than genotypes to be examined and found three TRDLs where there was a bias against S. aethnensis alleles and one TRDL where there was a bias against S. chrysanthemifolius alleles. Thus, pre-zygotic factors of the type mentioned above could have contributed to the TRD recorded at these loci.
Incompatibility between diverging genomes is often asymmetric when the underlying causes of incompatibility include either pre-zygotic haploid stages of the life cycle, such as meiotic drive or pollen fitness, or cytonuclear incompatibilities between nuclear and organelle genomes (Levin, 2003; Fishman and Willis, 2006; Turelli and Moyle, 2007). Strong crossing asymmetry or unilateral incompatibility is most likely observed when few asymmetric incompatibilities of large effect are involved (Turelli and Moyle, 2007). The extent to which TRD was asymmetric and dependent on cross direction was tested by comparing TRD across loci in F2 progeny possessing cytoplasm inherited from either the S. aethnensis or S. chrysanthemifolius parent. These tests revealed five TRDLs exhibiting asymmetric differences in TRD dependent on parental cytoplasmic background. We may conclude that the asymmetric TRD at these loci reflects cytonuclear incompatibility and/or the effects of meiotic drive and/or gametophytic selection.
There is growing evidence for the widespread occurrence of BDM incompatibilities between allopatric or parapatric populations and their role in limiting subsequent hybridization and possibly promoting further divergence during speciation (Coyne and Orr, 2004; Corbett-Detig et al., 2013). These BDM incompatibilities are frequently caused by deleterious interactions between different parental alleles occurring at two or more loci. Significantly non-independent, paired-locus genotype frequencies were observed for five pairs of TRDLs affecting four of the nine TRDLs (Table 3). However, three of the five interacting TRDL pairs were present in the same linkage group indicating that physical proximity might contribute to their non-independence. The under-represented genotype combination typically included one of the loci homozygous for S. aethnensis alleles and the other locus homozygous for S. chrysanthemifolius alleles.
We also tested if TRD could be due to (i) underdominance (reflected by a deficiency of heterozygotes at certain loci) or (ii) inbreeding depression, heterosis or recessive BDM incompatibilities (reflected by an excess of heterozygotes). Inbreeding depression could be caused by the expression of deleterious recessive alleles as a result of using a full-sib F2 mapping family from two outcrossed self-incompatible parents. Tests of heterozygosity revealed that underdominance could have contributed to a significant deficiency of heterozygous genotypes at only one TRDL, in the AC1 linkage group. However, the heterozygote deficiency at this locus was more likely caused by selection against S. aethnensis alleles, rather than underdominance, because there was an even stronger bias against genotypes homozygous for S. aethnensis alleles across the AC1 TRDL (Supplementary Figure S1). An excess of heterozygotes was not evident at any TRDL and thus it is concluded that inbreeding depression, heterosis or recessive BDM incompatibilities have not contributed to the TRD recorded in the F2AC mapping family.
As mentioned earlier in the discussion, reduced recombination near centromeres or due to local inversions may be causes of marker clustering observed in the genetic map. These causes of reduced recombination may also contribute to TRD at marker loci. For example, selfish drive elements are typically found near centromeres where they directly influence chromosome segregation patterns in their favour during meiosis (Henikoff et al., 2001), whereas chromosomal rearrangements may also influence inheritance patterns because recombination within the rearranged region is typically selected against (Ortiz-Barrientos et al., 2002, Kirkpatrick and Barton, 2006; Lowry and Willis, 2010). To examine whether genomic regions showing limited recombination could be a cause of TRD in the F2AC family, we tested for, but did not find associations between, marker clustering and TRDLs. Therefore, genomic regions showing limited recombination do not seem to have a strong role in reinforcing genomic divergence in Senecio. However, our current F2 genetic map lacks sufficient marker density to adequately test this association, as the clumped marker map distribution could be due to a variety of reasons other than variation in recombination rates and the genomic scale of TRDL-low recombination associations could be highly localized (Yatabe et al., 2007; Jones et al., 2012; Renaut et al., 2013). Future studies aimed at determining whether differences in chromosomal rearrangement may be a cause of genomic incompatibility between S. aethnensis and S. chrysanthemifolius should involve cytogenetic comparisons of karyotypes and/or detailed comparisons of high marker-density genetic maps of the two species.
Overall, it is clear from our analyses that a variety of genetic mechanisms at multiple genetic loci across the genome are likely to contribute to the intrinsic incompatibility existing between S. aethnensis and S. chrysanthemifolius. Further fine-scale genetic mapping studies of S. aethnensis and S. chrysanthemifolius involving quantitative trait locus mapping of traits of adaptive relevance, larger families to better estimate recombination, more markers for better genome coverage, and families from intraspecific crosses, will be necessary to further investigate the various mechanisms contributing to hybrid incompatibility and ecological divergence in this species pair.
Conclusions
While a number of studies have indicated that S. aethnensis and S. chrysanthemifolius are highly interfertile (Chapman et al., 2005; Brennan et al., 2013), the present study has revealed evidence of intrinsic incompatibility between these two species in the form of multiple genomic regions showing TRD. Our results, therefore, support the findings of our previous clinal analysis of the natural hybrid zone on Mount Etna, which indicated that intrinsic selection against hybrids was an important factor maintaining species differences in the face of gene flow (Brennan et al., 2009). The present study shows that hybrid incompatibility between these two diverging plant lineages is more cryptic than previously considered, and is manifested in the F2 generation rather than in the F1 (Hegarty et al., 2009). Some studies of other diverging plant lineages have yielded similar findings (Fishman et al., 2001; Moyle and Graham, 2006; Fishman and Willis, 2006).
It follows that the effects of TRDLs with multiple modes of action at multiple, relatively large, genomic regions in early-generation hybrids between S. aethnensis and S. chrysanthemifolius are likely to impact the genetic structure of the natural hybrid zone on Mount Etna by limiting introgression and promoting divergence across the genome (Feder et al., 2012; Abbott et al., 2013). However, large-scale genomic translocations or other rearrangements between the species do not seem to contribute in any major way to this process. It has been estimated that S. aethnensis and S. chrysanthemifolius are of relative recent origin (<1 million years ago, Comes and Abbott, 2001; ∼108 000 to 150 000 years ago, Chapman et al., 2013, Osborne et al., 2013), which indicates that the various forms of intrinsic hybrid incompatibility that clearly exist between these two species must have evolved relatively rapidly.
Data archiving
Mapping family genotype data available from the Dryad Digital Repository: doi:10.5061/dryad.7b56k. Other results can be found in Supplementary Information.
Acknowledgments
We thank David Forbes for technical assistance, Daniel Barker, Guo-Qing Liu and Ai-Lan Wang for help with the development of molecular markers and Lila Fishman and several anonymous referees for constructive comments on earlier drafts of the manuscript. The research was funded by a NERC Grant NE/D014166/1 to RJA as Principal Investigator. ACB was supported during the writing of this paper by funding from FP7-REGPOT 2010-1, Grant No. 264125 EcoGenes.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N et al. (2013). Hybridization and speciation. J Evol Biol 26: 229–246. [DOI] [PubMed] [Google Scholar]
- Abbott RJ, Brennan AC, James JK, Forbes DF, Hegarty MJ, Hiscock SJ. (2009). Recent hybrid origin and invasion of the British Isles by a self-incompatible species, Oxford ragwort (Senecio squalidus L., Asteraceae). Biol Invasions 11: 1145–1158. [Google Scholar]
- Agrawal AF, Feder JL, Nosil P. (2011). Ecological divergence and the origin of intrinsic postmating isolation with gene flow. Int J Ecol 2011: 435357. [Google Scholar]
- Alexander JCM. (1979). Mediterranean species of Senecio sections Senecio and Delphinifolius. Notes Roy Bot Gard Edin 37: 387–428. [Google Scholar]
- Bierne N, Welch J, Loire E, Bonhomme F, David P. (2011). The coupling hypothesis: why genome scans may fail to map local adaptation genes. Mol Ecol 20: 2044–2072. [DOI] [PubMed] [Google Scholar]
- Bomblies K. (2013). Genes causing postzygotic hybrid incompatibility in plants: a window into co-evolution. In: Chen ZJ, Birchler JA (eds) Polyploid and Hybrid Genomics. John Wiley & Sons, Inc.: Oxford, UK. pp 225–240. [Google Scholar]
- Brennan AC, Bridle JR, Wang A-L, Hiscock SJ, Abbott RJ. (2009). Adaptation and selection in the Senecio (Asteraceae) hybrid zone on Mount Etna, Sicily. New Phytol 183: 702–717. [DOI] [PubMed] [Google Scholar]
- Brennan AC, Harris SA, Hiscock SJ. (2013). The population genetics of sporophytic self-incompatibility in three hybridizing Senecio (Asteraceae) species with contrasting population histories. Evolution 67: 1347–1367. [DOI] [PubMed] [Google Scholar]
- Chakravarti A, Lasher LK, Reefer JE. (1991). A maximum likelihood method for estimating genome length using genetic linkage data. Genetics 128: 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Chang J, Weisman D, Kesseli RV, Burke JM. (2007). Universal markers for comparative mapping and phylogenetic analysis in the Asteraceae (Compositae). Theor Appl Genet 115: 747–755. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Forbes DG, Abbott RJ. (2005). Pollen competition among two species of Senecio (Asteraceae) that form a hybrid zone on Mt. Etna, Sicily. Am J Bot 92: 730–735. [DOI] [PubMed] [Google Scholar]
- Chapman MA, Hiscock SJ, Filatov DA. (2013). Genomic divergence during speciation driven by adaptation to altitude. Mol Biol Evol 30: 2553–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comes HP, Abbott RJ. (2001). Molecular phylogeography, reticulation and lineage sorting in the Mediterranean species complex of Senecio sect. Senecio (Asteraceae). Evolution 55: 1943–1962. [PubMed] [Google Scholar]
- Corbett-Detig R, Zhou J, Clark AG, Hartl DL, Ayroles JF. (2013). Genetic incompatibilities are widespread within species. Nature 504: 135–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. (2004) Speciation. Sinauer Associates: Sunderland, MA, USA. [Google Scholar]
- Cutter AD. (2012). The polymorphic prelude to Bateson-Dobzhansky-Muller incompatibilities. Trends Ecol Evol 27: 209–218. [DOI] [PubMed] [Google Scholar]
- Feder JL, Egan SP, Nosil P. (2012). The genomics of speciation with gene flow. Trends Genet 28: 342–350. [DOI] [PubMed] [Google Scholar]
- Fishman L, Aagaard J, Tuthill JC. (2008). Toward the evolutionary genomics of gametophytic divergence: patterns of transmission ratio distortion in monkeyflower (Mimulus) hybrids reveal a complex genetic basis for conspecific pollen precedence. Evolution 62: 2958–2970. [DOI] [PubMed] [Google Scholar]
- Fishman L, Kelly AJ, Morgan E, Willis JH. (2001). A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159: 1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Stathos A, Beardsley PM, Williams CF, Hill JP. (2013). Chromosomal rearrangements and the genetics of reproductive barriers in Mimulus (monkeyflowers). Evolution 69: 2547–2560. [DOI] [PubMed] [Google Scholar]
- Fishman L, Willis JH. (2005). A novel meiotic drive locus almost completely distorts segregation in Mimulus (monkeyflower) hybrids. Genetics 169: 347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman L, Willis JH. (2006). A cytonuclear incompatibility causes anther sterility in Mimulus hybrids. Evolution 60: 1372–1381. [DOI] [PubMed] [Google Scholar]
- Hegarty MJ, Barker GL, Brennan AC, Edwards KJ, Abbott RJ, Hiscock SJ. (2008). Changes to gene expression associated with hybrid speciation in plants: further insights from transcriptomic studies in Senecio. Phil Trans R Soc B 363: 3055–3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegarty MJ, Barker GL, Brennan AC, Edwards KJ, Abbott RJ, Hiscock SJ. (2009). Extreme changes to gene expression associated with homoploid hybrid speciation. Mol Ecl 18: 877–889. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. (2001). Speciation and centromere evolution. Science 294: 2478–2480. [DOI] [PubMed] [Google Scholar]
- James JK, Abbott RJ. (2005). Recent, allopatric, homoploid hybrid speciation: the origin of Oxford ragwort, Senecio squalidus (Asteraceae), in the British Isles from a hybrid zone on Mount Etna, Sicily. Evolution 59: 2533–2547. [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J et al. (2012). The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484: 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Cui ML, Cubas P, Gillies A, Lee K, Chapman MA et al. (2008). Regulatory genes control a key morphological and ecological trait transferred between species. Science 322: 1116–1119. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M, Barton NH. (2006). Chromosome inversions, local adaptation and speciation. Genetics 173: 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z, Nakazato T, Salmaso M, Burke JM, Tang S, Knapp SJ, Rieseberg LH. (2005). Extensive chromosomal repatterning and the evolution of sterility barriers in hybrid sunflower species. Genetics 171: 291–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latta RG, Gardner KM, Johansen-Morris AD. (2007). Hybridization, recombination, and the genetic basis of fitness variation across environments in Avena barbata. Genetica 129: 167–177. [DOI] [PubMed] [Google Scholar]
- Le Roux JJ, Wieczorek AM. (2006). Isolation and characterization of polymorphic microsatellite markers from fireweed, Senecio madagascariensis Poir. (Asteraceae). Mol Ecol Notes 7: 327–329. [Google Scholar]
- Levin DA. (2003). The cytoplasmic factor in plant speciation. Syst Bot 28: 5–11. [Google Scholar]
- Levin DA. (2012). The long wait for hybrid sterility in flowering plants. New Phytol 196: 666–670. [DOI] [PubMed] [Google Scholar]
- Liu G-Q, Hegarty MJ, Edwards KJ, Hiscock SJ, Abbott RJ. (2004). Isolation and characterization of microsatellite loci in Senecio. Mol. Ecol. Notes 4: 611–614. [Google Scholar]
- Lowry DB, Willis JH. (2010). A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol 8: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Romero-Seveson J, Bernardo R. (2002). Chromosomal regions associated with segregation distortion in maize. Theor Appl Genet 105: 622–628. [DOI] [PubMed] [Google Scholar]
- Matute DR, Butler IA, Turissini DA, Coyne JA. (2010). A test of the snowball theory for the rate of evolution of hybrid incompatibilities. Science 329: 1518–1521. [DOI] [PubMed] [Google Scholar]
- McInnis SM, Costa LM, Gutiérrez-Marcos JF, Henderson CA, Hiscock SJ. (2005). Isolation and characterization of a polymorphic stigma-specific class III peroxidase gene from Senecio squalidus L. (Asteraceae). Plant Mol Biol 57: 659–677. [DOI] [PubMed] [Google Scholar]
- Moyle LC, Graham EB. (2006). Genome-wide associations between hybrid sterility QTL and marker transmission ratio distortion. Mol Biol Evol 23: 973–980. [DOI] [PubMed] [Google Scholar]
- Moyle LC, Nakazato T. (2010). Hybrid incompatibility ‘snowballs' between Solanum species. Science 329: 1521–1523. [DOI] [PubMed] [Google Scholar]
- Muir G, Osborne OG, Sarasa J, Hiscock SJ, Filatov DA. (2013). Recent ecological selection on regulatory divergence is shaping clinal variation in Senecio on Mount Etna. Evolution 67: 3032–3042. [DOI] [PubMed] [Google Scholar]
- Nosil P. (2012) Ecological Speciation. Oxford University Press: Oxford. [Google Scholar]
- Ortiz-Barrientos D, Reiland J, Hey J, Noor MAF. (2002). Recombination and the divergence of hybridizing species. Genetica 116: 167–178. [PubMed] [Google Scholar]
- Osborne OG, Batstone TE, Hiscock SJ, Filatov DA. (2013). Rapid speciation with gene flow following the formation of Mt. Etna. Genome Biol Evol 5: 1704–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Zhang Q. (2013). Understanding reproductive isolation based on the rice model. Ann Rev Plant Biol 64: 11–35. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. (2011) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna. [Google Scholar]
- Remington DL, O'Malley DM. (2000). Whole genome characterization of embryonic stage inbreeding depression in a selfed Loblolly pine family. Genetics 155: 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaut S, Grassa CJ, Yeaman S, Moyers BT, Lai Z, Kane NC et al. (2013). Genomic islands of divergence are not affected by geography of speciation in sunflowers. Nat Commun 4: 1827. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. (2001). Chromosomal rearrangements and speciation. Trends Ecol Evol 16: 351–358. [DOI] [PubMed] [Google Scholar]
- Ross RIC, Ågren JA, Pannell JR. (2012). Exogenous selection shapes germination behaviour and seedling traits of populations at different altitudes in a Senecio hybrid zone. Ann Bot 110: 1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundle HD, Nosil P. (2005). Ecological speciation. Ecol Lett 8: 336–352. [Google Scholar]
- Schwarz-Sommer Z, de Andrade Silva E, Berndtgen R, Lönnig WE, Müller A, Nindl I et al. (2003). A linkage map of an F2 hybrid population of Antirrhinum majus and A. molle. Genetics 163: 699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemshorn KC, Reed FA, Nolte AW, Tautz D. (2011). Rapid formation of distinct hybrid lineages after secondary contact of two fish species (Cottus sp.). Mol Ecol 20: 1475–1491. [DOI] [PubMed] [Google Scholar]
- Tang S, Okashah RA, Knapp SJ, Arnold ML, Martin NH. (2010). Transmission ratio distortion results in asymmetric introgression in Louisiana Iris. BMC Plant Biol 10: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SJ, Rojas LD, Ho SW, Martin NH. (2012). Genomic collinearity and the genetic architecture of floral differences between the homoploid hybrid species Iris nelsonii and one of its progenitors, Iris hexagona. Heredity 110: 63–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Moyle LC. (2007). Asymmetric postmating isolation: Darwin's corollary to Haldane's rule. Genetics 176: 1059–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW. (2001) JoinMap v3.0 Software for the calculation of genetic linkage maps. Plant Research International: Wageningen, the Netherlands http://www.joinmap.nl.
- Voorrips RE. (2002). MapChart: Software for the graphical presentation of linkage maps and QTLs. J Hered 93: 77–78. [DOI] [PubMed] [Google Scholar]
- Xu S. (2008). Quantitative trait locus mapping can benefit from segregation distortion. Genetics 180: 2201–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatabe Y, Kane NC, Scotti-Saintaigne C, Reisberg LH. (2007). Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics 175: 1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.