Figure 1.
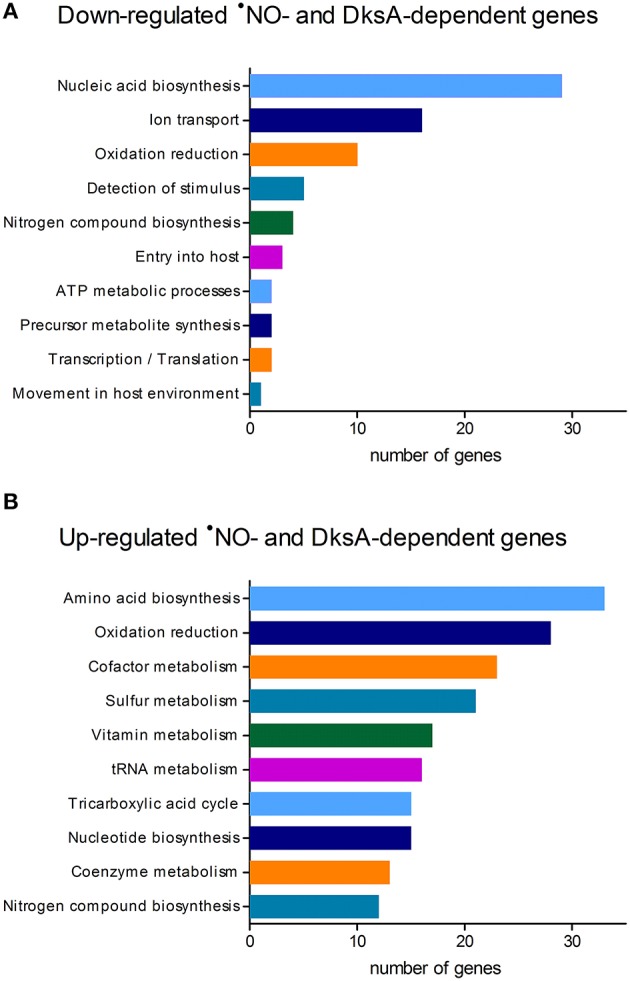
Functional annotation of •NO- and DksA-dependent genes in Salmonella. Genome-wide expression analysis of wild-type and ΔdksA S. enterica serovar Typhimurium strain 14028s treated ±5 mM dNO, for 30 min at 37°C with continuous shaking, identified 427 genes (~12% of all annotated open reading frames) as regulated in an •NO- and DksA-dependent manner. Microarrays were performed with cDNA generated from three independent experiments. Distribution of DksA-dependent genes (A) down-regulated or (B) up-regulated in wild-type Salmonella experiencing nitrosative stress; the top 10 categories are shown for each. Functional annotations were determined using Gene Ontology relationships.
