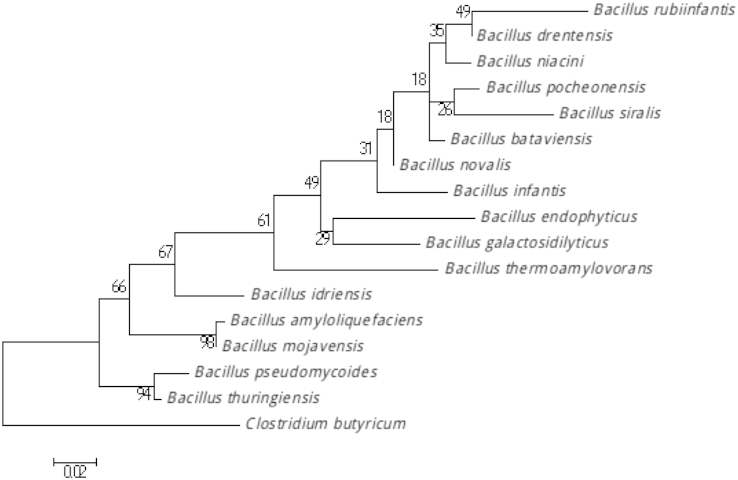
Fig. 1.
Phylogenetic tree highlighting position of Bacillus rubiinfantis sp. nov. strain mt2T (= CSUR P1141 = DSM 28615) relative to other type strains within Bacillus genus. Strains and their corresponding GenBank accession numbers for 16S rRNA genes are (type = T): B. novalis strain SCTB 113, JN650278; B. drentensis DRG 4, HQ436340; B. niacini strain TSII-13, JN993716; B. bataviensis strain LMG21832, AJ542507; B. pocheonensis strain Gsoil 420, AB245377.1; B. infantis strain A-49, KC751070; B. siralis strain 171544, NR_028709.1; B. endophyticus strain 2DT, NR_025122.1; B. pseudomycoides strain BIHB 360, FJ859700.1; B. thuringiensis strain GMC 108, AB741470; B. idriensis strain SMC 4352–2, NR_043268.1; B. amyloliquefaciens strain BIHB 35, FJ859496 ; B. mojavensis strain BCRC 17124, EF433405.1 ; B. galactosidilyticus strain LMG 17892, NR_025580.1 ; B. thermoamylovorans strain DKP, NR_029151.1. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA6. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 1000 times to generate majority consensus tree. Bacteroides thetaiotaomicron strain ATCC 29148T (L16489) was used as outgroup. Scale bar = 1% nucleotide sequence divergence.