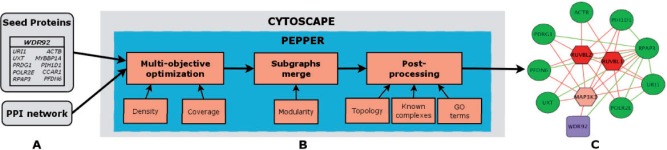
Fig. 1.
Schematic representation of the plug-in. (A) Example of input data, a large-scale PPI and the results of an AP-MS experiment with the bait and a list of prey proteins. (B) Context-specific protein complex extraction pipeline. (C) Output subnetwork representing a putative protein complex using only interactions from the input PPI network: example of WDR92. Purple squares and green circles correspond to bait and prey proteins, respectively. Hexagons indicate the expansions proposed by PEPPER and are shown in various shades of red, according to their post-processing score. Dark red indicates a high predicted relevance to the solution. The edges shown in the graph are exclusively those found in the input PPI network. Green edges are set between seed proteins. All edges involving an expansion protein are red