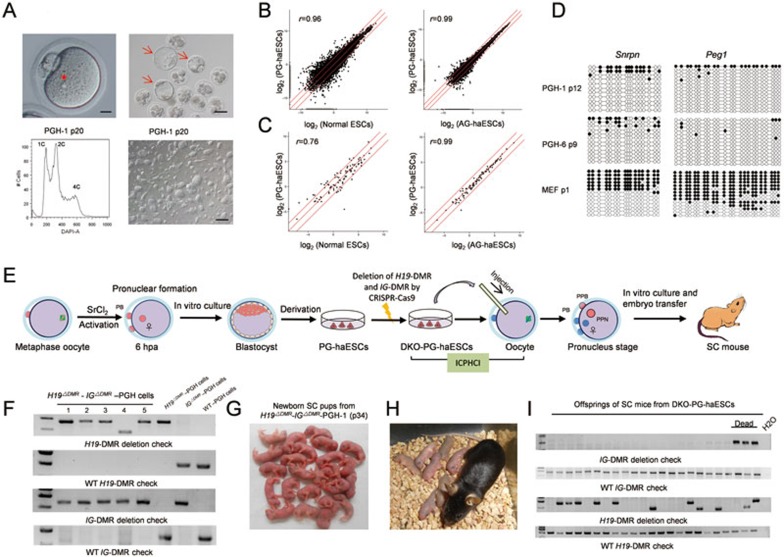
Figure 1.
PG-haESCs harboring both H19-DMR and IG-DMR deletions efficiently support the production of live SC pups. (A) Generation of PG-haESCs. Upper left, the image of parthenogenetically activated oocytes containing only one pronucleus (scale bar, 25 μm). Upper right, parthenogenetic blastocysts (scale bar, 100 μm). Lower left, establishment of a haploid cell line after multiple rounds of FACS enrichment for haploid cells (represented by PGH-1). A DAPI filter was used to detect the signal of Hoechst-stained DNA. Lower right, phase-contrast image of ESCs derived from one parthenogenetic blastocyst (scale bar, 200 μm). The asterisk indicates that only one pronucleus exists in the activated oocyte. Arrows indicate the parthenogenetic blastocysts that are used for ESC derivation. (B) Scatter plot of log2-transformed average gene expression profiles. Global gene expression profiles of PG-haESCs, AG-haESCs and diploid ESCs were obtained from RNA-seq analysis (r is the Pearson's correlation coefficient; red lines indicate twofold upregulation and downregulation). To avoid the influence of diploidized cells on the expression profile, we collected samples after FACS of cells at the G1/G0 phase. (C) Scatter plot analysis of gene expression profiles of the tested cell lines based on all imprinted genes. (D) Methylation states of the DMRs of Snrpn and Peg1 in two PG-haESC lines. Open and filled circles represent unmethylated and methylated CpG sites, respectively. (E) Diagram of SC mice generated by ICPHCI using PG-haESCs. hpa, hours post activation. PPN, pseudopronucleus formed from injected PG-haESC. PB, polar body. PPB, pseudopolar body. (F) Genotyping analysis of DKO-PG-haESCs. These cell lines were generated by deletion of both IG and H19 DMRs in WT PG-haESCs. Note that different sized bands can be observed in H19-DMR deletion analysis because using two sgRNAs to remove the 3.8-kb H19 DMR will result in cell lines carrying different sequences around the H19-DMR cleavage sites due to random DNA repair followed by CRISPR-Cas9-mediated cleavage. (G) SC pups generated by ICPHCI using DKO-PG-haESCs (represented by H19ΔDMR- IGΔDMR-PGH-1 cells (passage 34)). (H) A female SC mouse derived from DKO-PG-haESCs and its progeny. (I) Genotyping analysis of the progeny of SC mice derived from DKO-PG-haESCs. Pups carrying mutant IG-DMR or mutations in both H19 and IG DMRs died shortly after birth. Note that different sized bands are observed in H19-DMR deletion analysis because offsprings were born from two SC mothers produced from two DKO-PG-haESC lines carrying different sequences around the H19-DMR cleavage sites.