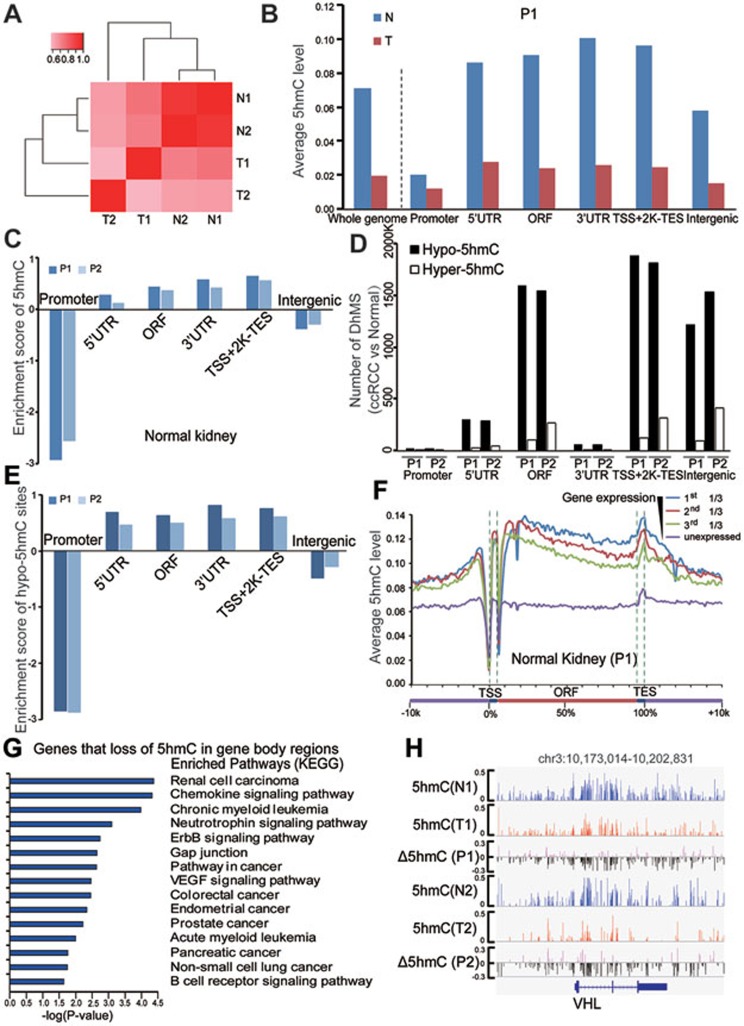
Figure 2.
Genome-wide single-nucleotide resolution mapping of 5hmC. (A) Correlations between 5hmC patterns with 5hmC level measured within 1 kb bins, and correlation coefficients are shown and colored from pink to red to indicate low to high. (B) Global changes of average 5hmC levels in different genomic elements determined by TAB-seq (promoter is defined as ± 500 bp of TSS). (C) The enrichment scores of called 5hmC sites in different genomic elements relative to expected. Score > 0 was defined as enriched. (D) The numbers of DhMS located in different gene-associated genetic elements. (E) The enrichment scores of hypo-5hmC sites in different genomic elements relative to expected. (F) The average 5hmC levels in normal kidney tissue across different gene-associated regions. Genes in the analyzed tissue were divided into four groups according to their gene expression levels (FPKM value). For each gene, a region from transcriptional start site (TSS) to transcriptional end site (TES) was divided into 100 bins and 5′-UTR and 3′-UTR contributed 5 bins each. Average 5hmC levels for each bin were calculated in the indicated samples. (G) The KEGG pathway analyses with the shared genes for both patients with loss of 5hmC in their gene body regions during tumorigenesis. The significance was evaluated by P-value. (H) Graphical representation of the dynamic 5hmC pattern during tumorigenesis at a 5hmC-enriched gene, VHL. T and N represent tumor and matched normal tissue, respectively. P1 and P2 represent patients 1 and 2, respectively.