Figure 2.
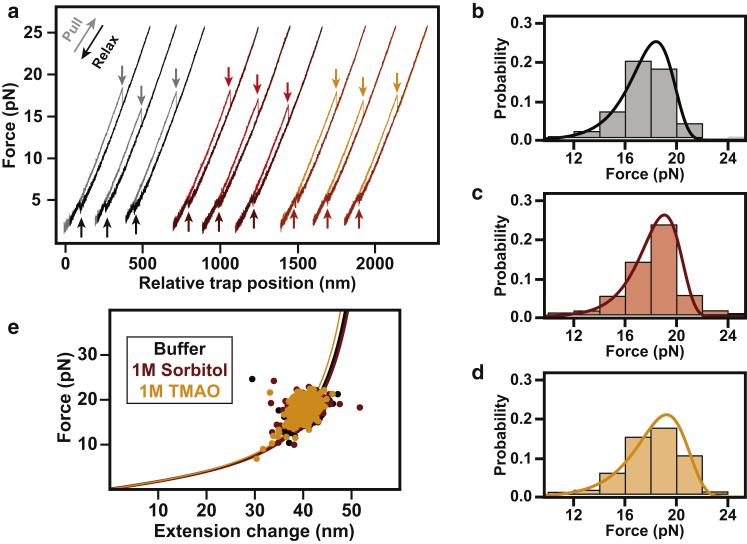
The mechanical unfolding pathway is unaffected by osmolyte (a). Shown are representative force-extension curves for single molecules of T4 lysozyme generated under constant velocity (100 nm/s). Force-extension curves in buffer (black), sorbitol (red), and TMAO (orange) behave similarly. Unfolding transitions (lighter arrows pointing down) were observed in the 12–24 pN regime for all conditions. Refolding occurred in the 3–6 pN regime (darker arrows pointing up). (b–d) Force rupture probability distributions in the presence of buffer (black), sorbitol (red), and TMAO (orange), respectively. Overlaid on the distributions are the fits using a theoretical model that yields distances to the transition state and lifetimes of the folded state (38). The distributions are well determined and the errors of the fits are smaller than the thickness of the lines. (e) Shown are the unfolding transition extension changes (in nanometers) versus the forces of unfolding (in picoNewtons) for every transition observed. Data are color-coded similar to (a) and are fit to the wormlike-chain model (55). All contour length changes (ΔLC) that are consistent with unfolding of the whole molecule are within error of each other. To see this figure in color, go online.