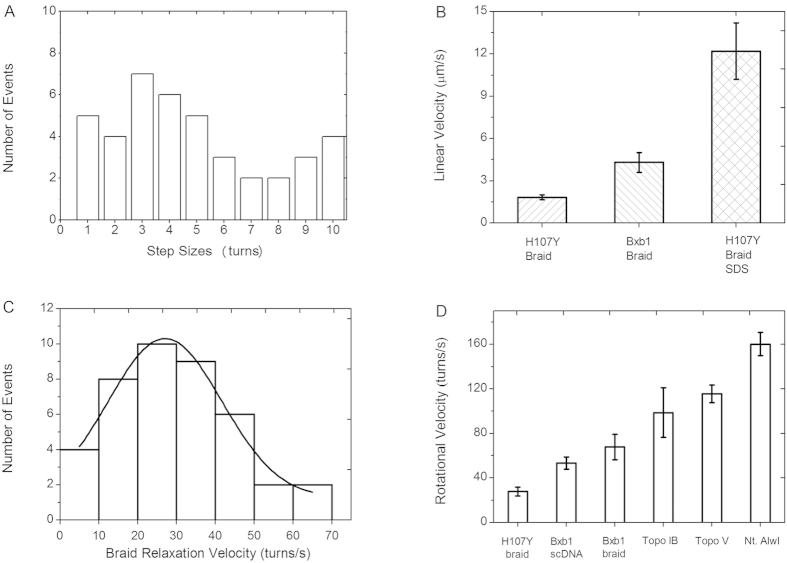
Figure 6. Event sizes and velocities for Hin-mediated braid relaxation.
(A) Distribution of relaxation event sizes. The number of events as a function of full rotations (360°) is plotted. The mean of the rotation distribution is 4.9 turns. (B) Comparison of linear velocities of Hin-H107Y braid relaxation (left bar) with release of the bead by SDS-triggered dissociation of Hin-H107Y DNA-cleaved complexes (right bar). Bead velocity for Bxb1 integrase (middle bar26) is shown for comparison. (C) Rotational velocities from the 41 analyzed relaxation steps by Hin-H107Y obeyed a Gaussian distribution with a mean of 27 turns/s and a SD of 14.6; the maximum velocity observed was 69.4 turns/s. (D) Comparison of mean rotational velocities: Hin-H107Y braid relaxation, Bxb1 serine integrase supercoil relaxation, Bxb1 serine integrase braid relaxation, Topo IB supercoil relaxation, Topo IV supercoil relaxation, Nt. AlwI supercoil relaxation (data for enzymes other than Hin from26).