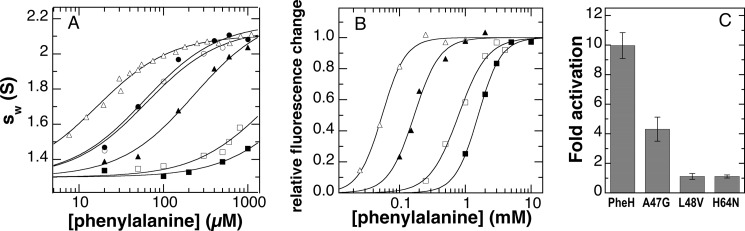
FIGURE 6.
Phenylalanine binding and activation of PheH variants. A, effect of the concentration of phenylalanine on the weight-average sedimentation coefficient (sw) of wild type (▵), L62V (○), E44Q (●), A47G (▴), H64N (□), and L48V (■) RDPheH(25–117). The lines are from fits of the data to sw = sm + Δsw × [Phe]/(KPhe + [Phe]). The value of sm was set to 1.3 based on the sw values in the absence of phenylalanine. For H64N and L48V RDPheH(25–117), the sw value of the dimer was assumed to be identical to that for wild-type RDPheH(25–117). The data for wild-type RDPheH(25–117) are from Ref. 25. B, representative fluorescence changes upon binding of phenylalanine to wild-type PheH (▵), A47G PheH (▴), H64N PheH (□), and L48V PheH (■) in 0.2 m HEPES, pH 7.5, at 25 °C. The lines are from fits of the data to Δfluorescence = ΔFlmax x× [Phe]n/(Kactn + [Phe]n). C, activation of PheH variants by phenylalanine. Each enzyme (25 μm) was incubated with 250 μm phenylalanine at 23 °C for 10 min before being diluted 100-fold into assay mix containing 1 mm phenylalanine and all other assay components, and the initial rate of tyrosine formation was determined.