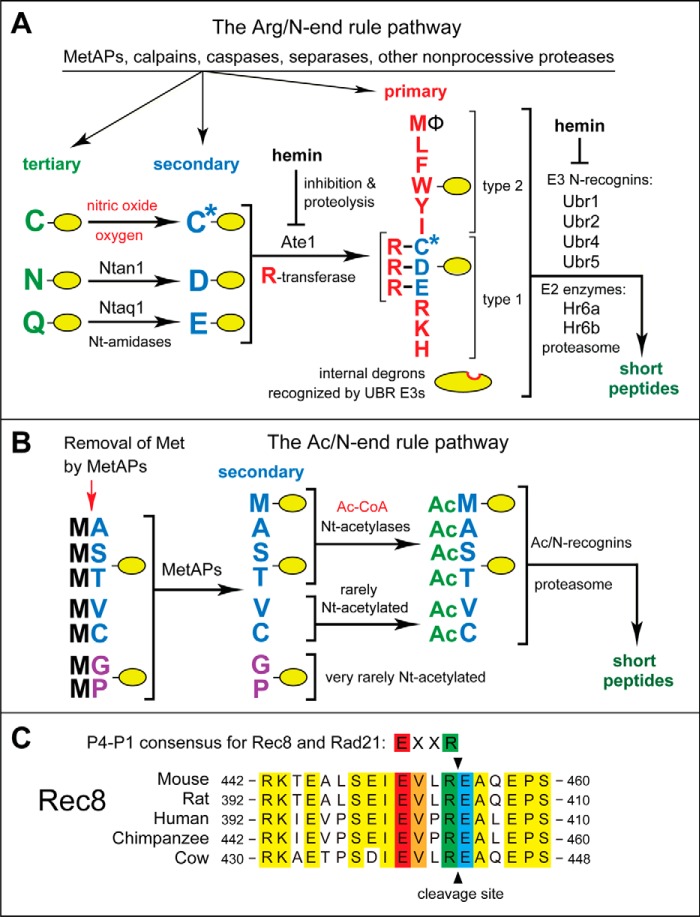
FIGURE 1.
The mammalian N-end rule pathway and the separase cleavage site in Rec8, a meiosis-specific cohesin subunit. See the Introduction for descriptions of the pathway's mechanistic aspects and biological functions. Amino acid residues are denoted by single-letter abbreviations. A, the Arg/N-end rule pathway. It targets proteins for degradation through their specific unacetylated N-terminal residues. A yellow oval denotes the rest of a protein substrate. R-transferase, Ate1 arginyltransferase; primary, secondary, and tertiary, mechanistically distinct classes of destabilizing N-terminal residues; type 1 and type 2, two sets of primary destabilizing N-terminal residues, basic (Arg, Lys, and His) and bulky hydrophobic (Leu, Phe, Trp, Tyr, Ile, and Met followed by a bulky hydrophobic residue (Φ)), respectively. These sets of N-terminal residues are recognized by two distinct substrate-binding sites of N-recognins, the pathway's E3 ubiquitin ligases. B, the Ac/N-end rule pathway. It targets proteins through their Nt-acetylated residues. The red arrow on the left indicates the cotranslational removal of the N-terminal Met residue by Met-aminopeptidases (MetAPs). N-terminal Met is retained if a residue at position 2 is larger than Val. C, alignments of amino acid sequences near the main separase cleavage site in mammalian Rec8, between Arg454 and Glu455 of mouse Rec8. The consensus sequence of this cleavage site (its P4–P1 residues) in both mitotic (Rad21) and meiotic (Rec8) kleisin-type cohesin subunits is also shown. Conserved residues of mammalian Rec8 near the cleavage site are shown in yellow. The conserved P4, P3, P1, and P1′ residues of mammalian Rec8 at the cleavage site are in red, orange, green, and blue, respectively.