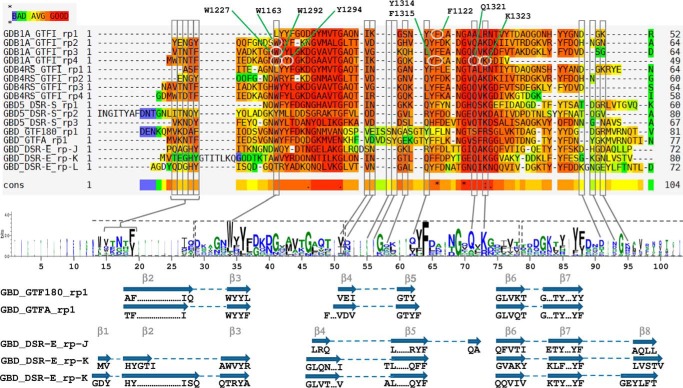
FIGURE 10.
Sequence alignment and LOGO sequence of the GBD repeats proved to bind α-glucans of GH70 glucansucrases. GBD1A, GBD4RS, and GBD5 bind dextrans with Kd values of 0.6 μm, 2.5 μm, and 11.1 nm, respectively. Sequences are GBD1A_GTFI_rp# (repeats 1 to 4 of the GBD1A of GTFI from S. downei), GBD-4RS_GTFI_rp# (repeats 1 to 4 of the GBD-4RS of GFTI from S. sobrinus), GBD5_DSR-S_rp# (repeats 1 to 3 of the GBD5 of DSR-S from L. mesenteroides B-512F), GBD_GTF180_rp1 (repeat 1 of the GBD of GTF180-ΔN from Lactobacillus reuteri), GBD_GTFA_rp1 (repeat 1 of the GBD of GTFA-ΔN from L. reuteri 121), and GBD_DSR-E_rp# (repeats J to L of the GBD of ΔN123-GBD-CD2 from L. citreum NRRL-1299). Residues highlighted in red are well conserved, whereas those tending toward blue are not. Residues framed in gray delineate the sugar binding pockets and are pointed out in the LOGO sequence. Residues framed in dashed line on the LOGO sequence correspond to YG repeats. In the repeats of GBD1A, residues circled in white, were mutated by Shah et al. (25) (see ”Discussion“). Secondary structure elements, as determined by DSSP from PDB entries 3TTQ, 3KLK, and 4AMC, are displayed below the LOGO sequence. β-Strands, numbered according to Fig. 8, linked by dashed lines represent either β-hairpins or three-stranded β-sheets.