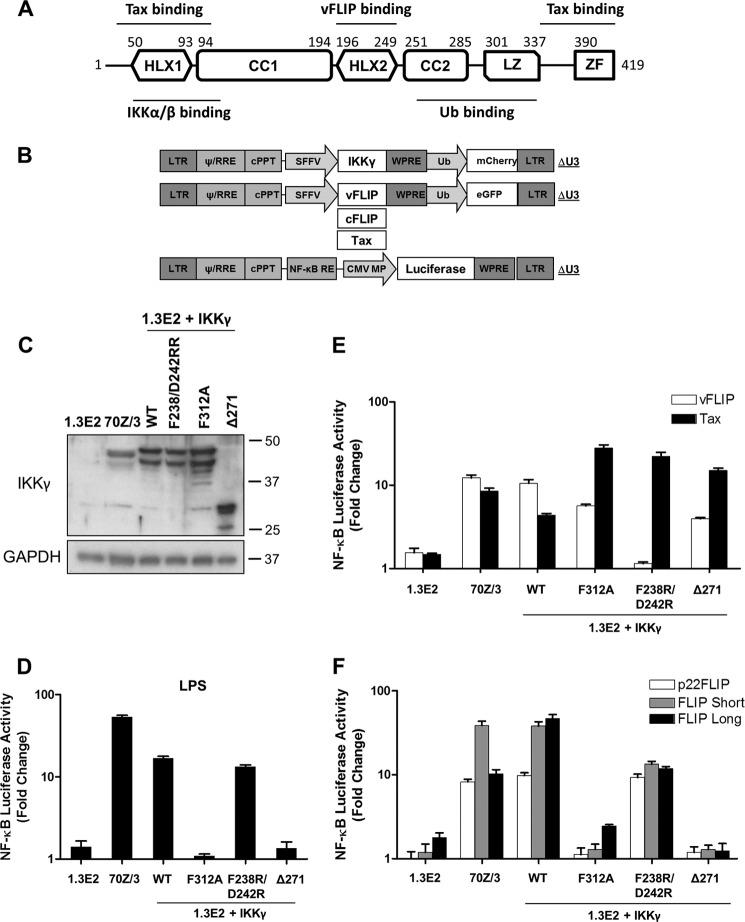
FIGURE 1.
Mutation of IKKγ reveals different NF-κB activation mechanisms by KSHV vFLIP and cellular FLIPs. A, schematic representation of IKKγ and the regions involved in interaction with IKKα/β, KSHV vFLIP, Tax, and ubiquitin (Ub). CC, coiled coil; HLX, helical domain; LZ, leucine zipper; ZF, zinc finger. B, map of the lentivectors. CMV MP, minimal promoter of cytomegalovirus; cPPT, central polypurine tract; LTR, long terminal repeat; NF-κB RE, NF-κB response element; RRE, rev response element; SFFV, spleen focus-forming virus; WPRE, woodchuck post-transcription regulatory element. C, immunoblot showing the expression of IKKγ in 1.3E2 cells reconstituted with wild-type or mutant IKKγ. The blot was reprobed for GAPDH. D, to generate stable NF-κB reporter cell lines, cells were transduced with lentivectors encoding an NF-κB-responsive luciferase gene. The luciferase reporter assays were performed 6 h after stimulation with LPS (10 μg/ml) or 48 h following transduction with lentivectors encoding vFLIP, Tax (E), and cFLIP variants (F). Data are representative of three independent experiments performed in triplicate. Error bars indicate S.D. of the mean values.