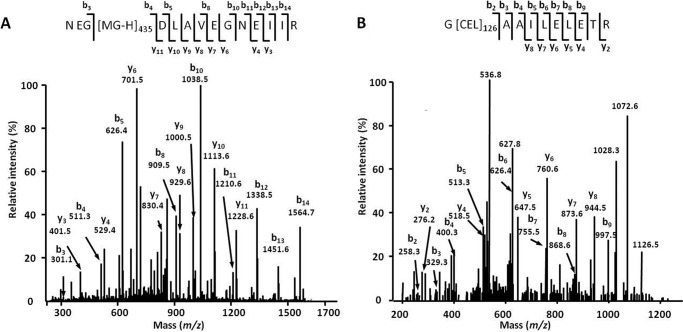
FIGURE 4.
Tandem mass spectra obtained for m/z 869. 94 (peptide NEG[MG-H]435DLAVEGNEIIR, large chain of Rubisco, EC 4.1.1.39; A) and m/z 636.86 (peptide G[CEL]126AAILELETR, enoyl-CoA hydratase 2, EC 4.2.1.119; B). The spectra were acquired by nanoscale UPLC-ESI-Orbitrap-LIT-MS operated in positive DDA mode. The peptide sequences were derived by database search and confirmed by manual interpretation. bn, N-terminal fragment ion series; yn, C-terminal fragment ion series; subscripts 435 and 126 represent the positions of MG-H and CEL in the corresponding polypeptide backbones.