Abstract
As a relatively recent research field, plant metabolomics has gained increasing interest in the past few years and has been applied to answer biological questions through large-scale qualitative and quantitative analyses of the plant metabolome. The combination of sensitivity and selectivity offered by mass spectrometry (MS) for measurement of many metabolites in a single shot makes it an indispensable platform in metabolomics. In this regard, Fourier-transform ion cyclotron resonance (FTICR) has the unique advantage of delivering high mass resolving power and mass accuracy simultaneously, making it ideal for the study of complex mixtures such as plant extracts. Here we optimize soybean leaf extraction methods compatible with high-throughput reproducible MS-based metabolomics. In addition, matrix-assisted laser desorption ionization (MALDI) and direct LDI of soybean leaves are compared for metabolite profiling. The extraction method combined with electrospray (ESI)-FTICR is supported by the significant reduction of chlorophyll and its related metabolites as the growing season moves from midsummer to the autumn harvest day. To our knowledge for the first time, the use of ESI-FTICR MS and MALDI-FTICR MS is described in a complementary manner with the aim of metabolic profiling of plant leaves that have been collected at different time points during the growing season.
Graphical Abstract
Metabolomics principally aims to measure and study small molecule metabolites that constitute the biological network as a means to better understand the precise biological role or function of a biological system at a defined developmental stage or under specific environmental factors1 such as pathological states of human diseases like cancer,2 diabetes,3 autoimmune,4 and coronary diseases.5 Because of the benefit produced on human health of various products of plant origin such as pharmaceuticals, food, and industrial raw materials, metabolomics is currently being extensively applied in plant research as well as in plant breeding and nutrition assessment. Owing to being highly dynamic in time and space and showing an immense range of structural variations, no single analytical tool encompassing the whole metabolome is currently available. Thus, numerous analytical procedures have been developed6 to provide both the needed selectivity and sensitivity for their analysis in highly complex mixtures. These include mass spectrometry (MS),7,8 nuclear magnetic resonance (NMR),9,10 and vibrational (IR or Raman) spectroscopy11,12 coupled with separation techniques, including gas chromatography (GC),13 liquid chromatography (LC),14 and capillary electrophoresis (CE).15 In metabolomics research it is often the case using various techniques simultaneously to obtain complementary information for the identification and/or quantitation of metabolites.
Among the several analytical platforms nuclear magnetic resonance spectroscopy (NMR) and mass spectrometry (MS) are considered to be the primary analytical technologies of metabolomics, where they have a great potential to complement each other. Inherently high sensitivity and selectivity provided by mass spectrometry makes it an increasingly important tool for the analysis of biological systems. Thus, in attempts to obtain better metabolite identification and quantitation, several hyphenated approaches such as GC/MS, LC/MS, and CE/MS, have been developed.16–19 There is no doubt that a significant breakthrough achieved in the field of MS-based metabolomics was introduction of novel soft ionization methods, namely, electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI), for mass spectrometric (MS) analysis of a large variety of biological molecules ranging from small metabolites to large proteins were facilitated. In combination with ESI and MALDI, mass spectrometry proved to be useful in the investigation of biological systems.20–23 Although, the plant tissues had to be extracted by solvents through laborious procedures, the decrease in the analysis time due to no requirement of up-front separation steps (adding more challenges in data processing due to the run-to-run variability of the chromatographic or electrophoretic separations) motivated scientists to investigate the primary and secondary plant metabolites by ESI-MS.24–26
The efforts on identification and absolute quantitation of human plasma metabolites revealed that Fourier-transform ion cyclotron resonance mass spectrometry (FTICR MS) has the potential to be a powerful new technique for high-throughput metabolomics as this technique delivers unmatched mass resolution, mass accuracy, and exceptional detection sensitivity.27 Moreover, often it requires no prior wet chemical separation, thereby potentially changing the whole approach to dealing with chemical and biological complexity.28
Herein we describe a novel analytical protocol, which consists of optimization of plant tissue extractions compatible with reproducible, high-throughput MS-based metabolomics and a combination of various soft ionization methods with FTICR MS technology for investigation of a plant metabolome. As a test material we have chosen soybean leaves. This is because, together with rice and corn, soybeans are among the most valuable cash crops in the world. Soybeans are used in a variety of products, primarily as protein and oil sources for humans, feed for livestock and aquaculture, and as a feedstock in biofuel production. Moreover, some of the chemical constituents in soybeans have been suggested to exhibit hypocholesterolemic, immunostimulatory, anticarcinogenic, antioxidatative, antidiabetic, and hepatoporotective activities, making the high-throughput profiling of soybean metabolome extremely important.29–31
EXPERIMENTAL SECTION
Chemicals
Methylene chloride (ACS grade) and methanol (HPLC grade) were from Fisher Scientific. Ethanol (200 proof) was from Decon Laboratories, Inc. Acetonitrile (HPLC grade) and bilirubin were from Sigma-Aldrich. Ammonium hydroxide and water (LC/MS grade) were from J. T. Baker. Alpha-4-cyanohydroxy cinnamic acid (CHCA) was from Fluka. Trifluoroacetic acid (TFA) was from Aldrich. Stercobilin hydrochloride was from Frontier Scientific (Logan, UT).
Samples Used during the Study
Soybean (Glycine max) leaves were harvested from a farm in Northern Indiana (latitude 41.588 059, longitude –86.211 358) at the middle and conclusion (harvesting day) of a growing season. The harvested leaves were immediately placed in a freezer bag and stored on ice and remained frozen until sample processing. For laser desorption and MALDI experiments, small whole leaves or leaf sections were mounted onto conductive indium–tin oxide (ITO) coated slides (Bruker Daltonics) using Mount Quick (Ted Pella, Inc.) glue; curing of the glue was allowed to proceed at room temperature. For MALDI, CHCA was applied as the matrix using a Bruker Image Prep system.
Extraction Protocols
For positive ion mode, in an attempt to have an increased coverage of the soybean leaf metabolome, ground soybean leaves were extracted with ethanol, dichloromethane, and acetonitrile (1 mL), respectively, by maceration for 30 min at room temperature with two 5 min periods of sonication at the beginning and at the end of the extraction period. The mixtures were diluted to a total volume of 10 mL and filtered with a 0.2 mm syringe filter. Subsequently, aliquots of the supernatant were transferred to glass vials for further analysis. For negative ion mode, following the pulverization, soybean leaves were extracted with 1 mL of mixture of ethanol–ammonium hydroxide, dichloromethane–ammonium hydroxide, and acetonitrile–ammonium hydroxide in the ratio of 99:1, respectively. That is followed by application of exactly the same protocol described above.
Data Collection and Processing
All mass spectra were collected on a Bruker Daltonics 12 T SolariX FTICR mass spectrometer equipped with an ESI source and a SmartBeam II frequency-tripled (355 nm) Nd:YAG MALDI source. Both positive and negative ion mode analysis was conducted. For positive ion mode ESI, the solvent extracts after filtration were directly analyzed; for negative ion mode ESI, the sample extracts contained 1% ammonium hydroxide by volume. For direct laser desorption, analysis was conducted on leaves mounted on the ITO slides without additional treatment. For MALDI, leaves mounted onto ITO slides were coated with matrix using the ImagePrep system with saturated CHCA solutions dissolved in 3:7 acetonitrile/0.1% aqueous TFA. For FTICR, mass spectra were collected as 1 or 2 MWord data sets and displayed in magnitude-mode. For peak-picking within the data sets, all peaks with signal-to-noise ≥5 were found, producing hundreds of unique m/z peaks in each mass spectrum. This complexity is only in part due to the high chemical complexity of the soybean leaf extracts; it is also complicated due to the existence of isotopic envelopes for each chemical species. To decrease the complexity of isotopic contributions and/or multiple charge states (for ESI), the mass spectra were deconvoluted using the resolved-isotope deconvolution function in Bruker’s Data Analysis software to produce peak lists for each component; ethanol extracts contained far more detected components than dichloromethane or acetonitrile extracts, or by direct laser desorption/MALDI analysis of the leaves, so ethanolic extracts were further examined for metabolite changes during the growing cycle. In the experiments where the quantities of each metabolite relative to an internal standard (stercobilin) were performed, five replicates were acquired. Using the online Metaboanalyst 3.0 Web-based statistical tool,32 peak lists of m/z and absolute intensity were aligned (0.0025 m/z window) and normalized relative to stercobilin; fold-change in abundance was calculated and heat maps of metabolite differences between the summer and autumn harvest leaf extracts were generated in Metaboanalyst.
RESULTS AND DISCUSSION
ESI of Soybean Leaf Extracts
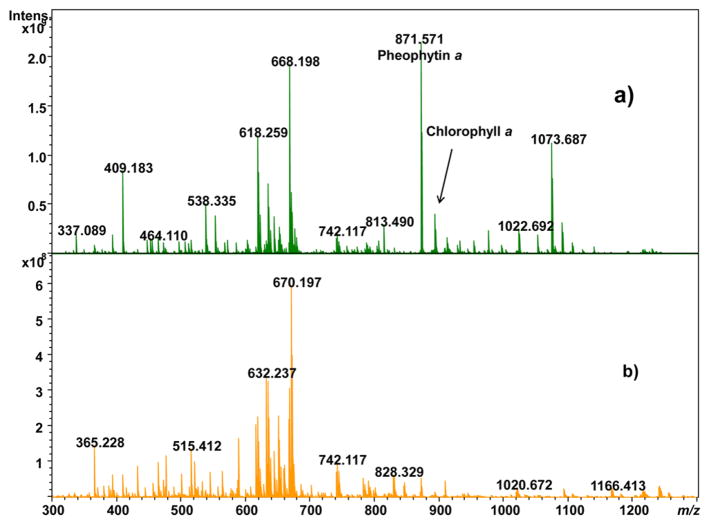
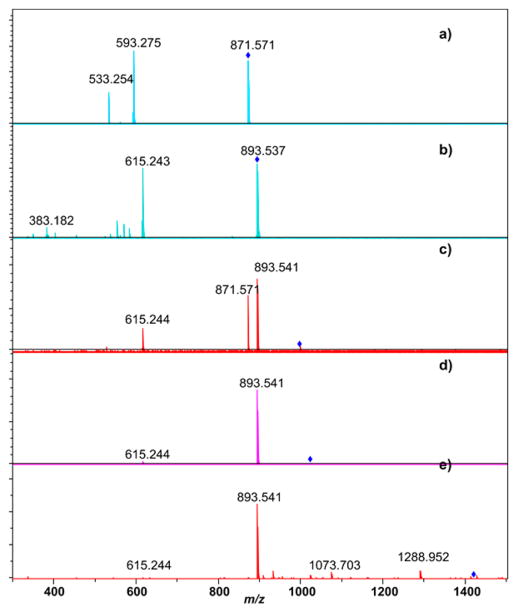
Soybean leaf extracts in ethanol (~330 different ion signals), acetonitrile (134 different ion signals, 80 of which were not observed in the ethanol extracts), and methylene chloride (36 different ion signals, 27 of which were not observed in the ethanol extracts) were performed and examined by positive and negative ion ESI-FTICR. Direct infusion ESI was chosen over LC-ESI to minimize method development time, storage size requirements of the data, analysis time, and cost. Extractions were performed on soybean leaves harvested in the middle of the growing season (midsummer) and on the final day of the growing season (autumn) immediately prior to harvest. For positive ion ESI, the mass spectra provided strong signal in the absence of any added acid or base. For negative ion ESI, optimal ion signal was observed when the extracts were spiked with 2% ammonium hydroxide. Extractions of the soybean leaves in ethanol provided significantly more ion signals than the extractions in acetonitrile or methylene chloride; a comparison of the ESI-FTICR mass spectra from ethanol extracts from summer-harvested leaves and autumn-harvested leaves is shown in Figure 1. Supplementary Figure 1 shows positive ion ESI-FTICR mass spectra of the other summer leaf extracts while Supplementary Figure 2 shows negative ion ESI-FTICR mass spectra of these extracts. There are notable differences in the appearance of ethanolic summer leaf extracts compared to autumn leaf extracts. The most intense peak observed in the ethanolic summer leaf extract in Figure 1a has an exact m/z 871.571. CID of this peak (Figure 2a) leads to loss of 278.296 amu, which corresponds to neutral loss of C20H38; such a neutral loss is known for chlorophylls including pheophytin a.33–36 Thus, the correspondence of exact mass to the (M + H)+ ion (−2.5 ppm) and the MS/MS data indicates this species is pheophytin a. The peak at m/z 893.540 corresponds to the (M + H)+ ion of chlorophyll a (−2.9 ppm), as confirmed by its CID tandem mass spectrum (Figure 2b); the (M + K)+ ion of chlorophyll a was also detected at m/z 931.496. Pheophytin a and chlorophyll a are still present in the autumn leaf extracts, but their abundances are significantly depleted (discussed in the following section). CID of three other metabolites at m/z 996.639, 1022.691, and 1418.098 (Figure 2c–e) indicate that these are closely related to chlorophyll a, as each produces a strong fragment ion consistent with chlorophyll a as well as a fragment due to loss of the phytyl group at m/z 615.2444. These substances do not match any previously identified chlorophyll metabolites, so isolation of these species and further NMR characterization to augment the MS/MS data is warranted. Weak intensity peaks are also observed for chlorophyll b for its (M + H)+ and (M + K)+ ions at m/z 907.519 and 945.475, respectively.
Figure 1.
Positive ion ESI-FTICR mass spectrum of ethanol extracts of soybean leaves: (a) midsummer-harvested and (b) autumn-harvested.
Figure 2.
ESI-FTICR MS/MS spectra of metabolites related to chlorophylls using collision induced dissociation: (a) pheophytin a, (b) chlorophyll a, (c) chlorophyll-related metabolite at m/z 996.639, (d) chlorophyll-related metabolite at m/z 1022.691, and (e) chlorophyll-related metabolite at m/z 1418.098.
Several peaks in the ethanol extracts of soybean leaves are attributed to the species dilauryl thiodipropionate with m/z 515.413, 537.394, and 553.368 corresponding to the (M + H)+ (+0.3 ppm), (M + Na)+ (−1.5 ppm error), and the (M + K)+ (−1.3 ppm) ions, respectively. The (M + 2Na – H)+ and (M + Na + K – H)+, and (2M + K)+ ions for this species are also detected, as are (M + H)+ and (2M + Na)+ and (2M + K)+ ions for a species oxidized to the sulfoxide derivative. Dilauryl thiodipropanoate is a common antioxidant found in plasticizers and cosmetics37 and has been detected as a leachate from polypropylene tubes38 but is not known to be endogenous to soybeans; therefore, these peaks were excluded in the analysis of metabolite changes according to growing season detailed below. Thus, while ethanol may effectively extract many components from soybean leaves, it is important for future studies to recognize potential contamination from dilauryl thiodipropionate onto leaf surfaces if specimens are stored in plastic bags used in food packaging; it may be advisible to use glass Petri dishes instead. Nevertheless, Figure 1 readily shows that the molecular composition of the extracts changes over the course of the growing season, the quantitative nature of which will be discussed in the following section.
Relative Quantitation Differences for Senescence from ESI-FTICR
As discussed above, pheophytin a and chlorophyll a, two key metabolites involved in photosynthesis, are prominent in ethanol extracts of summer-harvested leaves but are depleted in the autumn-harvested leaves. Many other m/z were decreased in autumn soy leaf extracts, while some m/ z are observed to increase as the season progresses from summer to autumn. Over 330 unique m/z were detected in both the summer and autumn extracts. In order to quantify the level of analyte change between seasons, 100 μL aliquots of ethanol extracts of leaf composites from both summer- and autumn-harvest were spiked with 2 μg of stercobilin for normalization. Stercobilin is a tetrapyrrole produced from the metabolism of heme in mammals that is additionally processed by microbiota in the colon; stercobilin is not known to exist in plants and is not observed in the unspiked ethanol leaf extracts. Because both chlorophyll a and stercobilin are structurally related to heme, and because of our own knowledge of its fragmentation behavior,39 it seemed an extremely appropriate reference for normalization in assessing the effect of seasonal variation in the abundance of key soybean leaf metabolites. The peak list generated was used to search the Human Metabolome Database (HMDB) for possible matches.40
Figure 3 displays a graphical representation of several m/z and log(fold change in normalized abundance) as a function of the time the soybean leaves were harvested; positive log(fold change) values therefore show greater abundance in summer, while those with negative log(fold change) values show greater abundance in autumn. Chlorophyll a is found to be greater by almost 300× in the summer-harvested ethanol leaf extracts in comparison to the autumn-harvested leaf extracts, consistent with the observation of substantially decreased green coloring of the autumn-harvested leaves. The depletion of pheophytin a is less dramatic, as its relative level falls by only 2.65×. Other metabolites we identified as being related to chlorophyll a are also more abundant in summer, with the m/z of 996.64 depleted 270× and m/z 1022.70 depleted 122×; chlorophyll b is depleted 120–132×. A full table of the metabolites and their relative fold change in abundance vs the internal standard in going from summer to autumn is provided as Supplementary Table 1; possible matches from HMDB are indicated in Supplementary Table 1. Other possible metabolites more than an order of magnitude more abundant in summer leaf extracts include soyacerebroside, several phospholipids, and numerous di- and triacyl glycerols.
Figure 3.
Plot of the log (fold change in relative intensity summer/ autumn) as a function of m/z for ethanol extracts of soybean leaves. All peak intensities have been normalized to the intensity of the stercobilin internal standard.
Likewise, the (M + Na)+ peak due to C40H56, which is attributed to carotenes, is only 6.1% the relative abundance level in summer as in the autumn leaf extracts. This does not necessarily mean that carotene content increased as the season progressed; it could be that the significant depletion of chlorophylls enabled more efficient ionization of carotene.
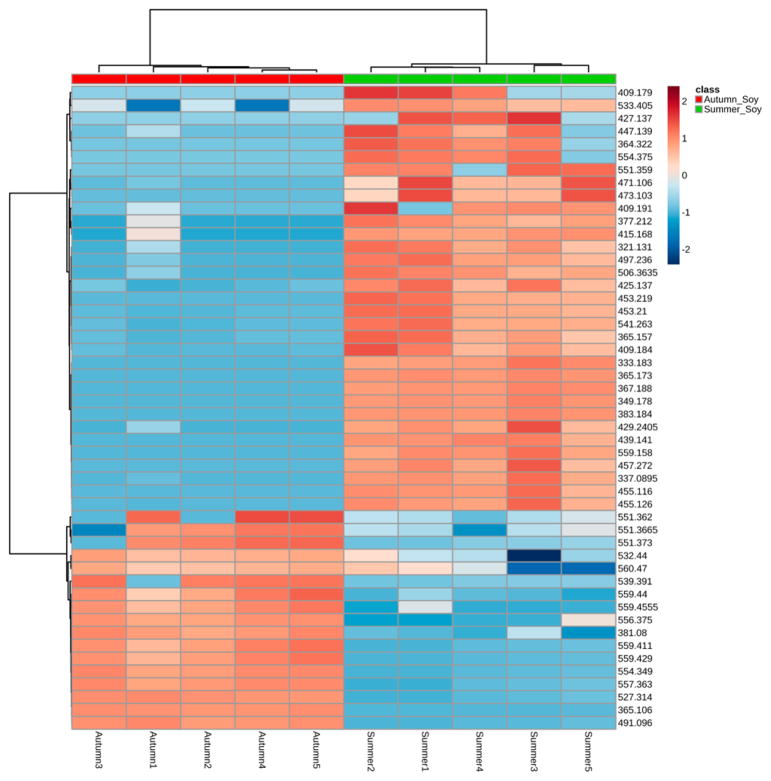
The disaccharides with the formula C12H22O11 are detected as (M + Na)+, which increases by almost a factor of 50 from summer to autumn, suggesting that the breakdown of complex carbohydrates is well under way in the autumn-harvested leaves. A heat map of the 50 most important metabolite differences in the summer and autumn leaf extracts generated by Metaboanalyst 3.0 is shown in Figure 4. The heat map very nicely demonstrates clear distinctions between the metabolite profiles and which metabolites are abundant in summer vs those which are abundant in autumn. However, it is important to recognize that the complexity of the leaf matrix may suppress ionization of some chemical constitutents; thus, even with normalization, caution needs to be exercised when quantitative analysis of leaves is performed.
Figure 4.
Heat map of relative intensity for replicates of ethanol extracts of soybean leaves harvested in summer and autumn. The heat map shows quite clearly that many metabolites deplete as the growing season progresses while others become much more abundant during the latter stages of the growing season.
MALDI-FTICR
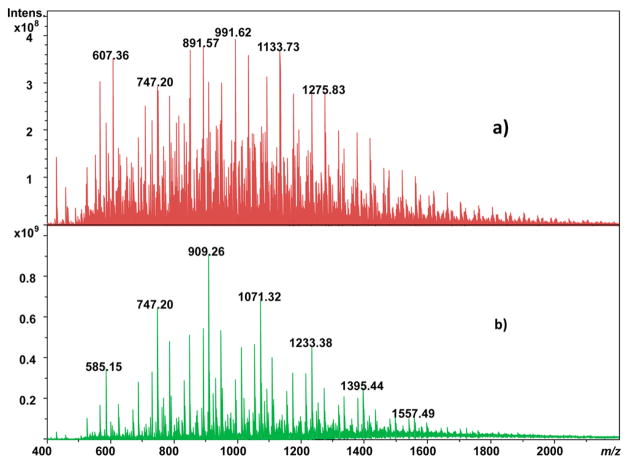
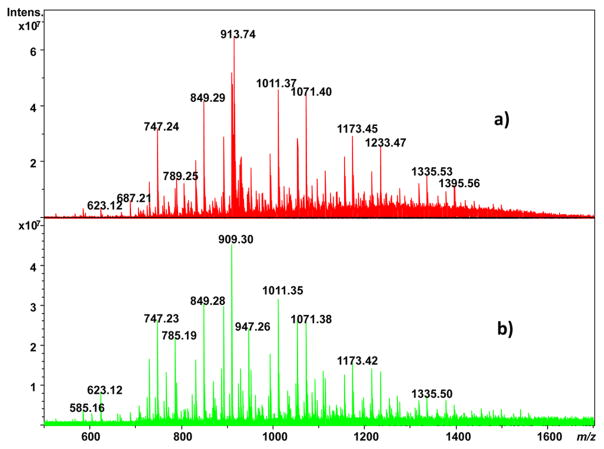
Another approach that has shown great promise for differentiation of seasonal variations in plant leaf metabolic profiles is leaf spray ionization.36 However, whether using ESI of solvent-extracted leaves or leaf spray, the use of solvents biases analysis to only those substances that are soluble in them. An approach that would avoid such bias and ionize substances directly from the leaf surface is MALDI. Here, MALDI is performed with CHCA matrix that is pneumatically deposited onto a soybean leaf affixed onto an ITO glass slide with Clear Mount glue (which is allowed to cure under ambient conditions). Seasonal variation in the metabolic profile of substances at the surface of soybean leaves is indicated in Figure 5; Figure 5a is the MALDI FTICR mass spectrum of a leaf collected on harvest day, while Figure 5b is the MALDI FTICR mass spectrum of a leaf collected during midsummer. While there are intensity differences between the ions observed, qualitatively most of the same m/z values are observed in both cases. Both the autumn- and summer-harvested leaves show a series of ions differing by an exact mass consistent with C2H4O2, for example, the pairs at 831.23/891.25 amu and 849.24/909.26 amu, respectively. Perhaps of more interest is that these species are not observed in the ethanol extracts of soybean leaves shown in Figure 1, indicating that MALDI can ionize compounds not solubilized in extracts amenable to ESI, highlighting the complementary information obtained by the MALDI and ESI approaches. Indeed, the ~320 unique signals of the extracts by MALDI, about 98% were not detected by ESI. Of these, 53 matched one (or more) candidate identifications with the HMDB database.
Figure 5.
MALDI-FTICR mass spectra of (a) autumn-harvested and (b) summer-harvested soybean leaf.
There are three striking differences between the MALDI mass spectra of the autumn- and summer-harvested leaves. First is the presence of a series of ions in Figure 5a differing by a mass consistent with C3H6; this series is absent in Figure 5b obtained from the summer-harvested leaves. In addition, the most prominent ion series in Figure 5b from the summer-harvested leaves exhibits a mass difference of 162.05 Da, which is consistent with hexose groups. This ion series is less prominent (although present) in the autumn-harvested leaves (e.g., the triad of peaks at 747.20, 909.26, and 1071.32 amu), especially at higher m/z. Likewise, there is a decrease in intensity of the related ion series shifted 60.02 Da lower than the hexose series, consistent with a difference in formula of C2H4O2.
LDI Comparison with MALDI
As noted above, MALDI showed differences between the metabolic profile whether harvested midsummer or autumn or based on different anatomic features in the leaf itself. MALDI expanded the applicability of laser desorption and ionization (LDI) methods by greatly increasing the mass range and substantially improving ion signal, even for lower molecular weight substances.41 However, it was unclear whether MALDI is absolutely necessary for profiling metabolites from soybean leaves. For example, Hölscher et al. demonstrated that matrix-free LDI can be performed for direct profiling of plant leaf and petal metabolites in imaging mass spectrometry because of the presence of UV-absorbing secondary metabolites.42 Thus, we performed direct LDI on the soybean leaf. The mass spectra generated by LDI on soybean leaves collected on the day of harvest and midsummer are compared in parts a and b of Figure 6, respectively. These mass spectra were collected in blade regions between veins. The direct LDI mass spectra are qualitatively very similar, whether the leaf was harvested in autumn or summer. This contrasts with the MALDI mass spectra of soybean leaves shown in Figure 5. Of the ~100 unique peaks, approximately 70% are unique to LDI, with only 5% overlap with ESI. Of these, only 16 matched one (or more) candidate identifications with the HMDB database. In addition, the absolute intensities of the detected ions are an order of magnitude smaller for LDI (absolute intensity ~107) vs MALDI (absolute intensity ~108). Therefore, it is clear that MALDI is preferable to LDI for soybean leaf metabolomics profiling. Supplementary Table 2 and Supplementary Table 3 provide lists of the monoisotopic m/z values for ions identified after deconvolution for MALDI and LDI, respectively.
Figure 6.
Direct LDI-FTICR mass spectra of (a) autumn-harvested and (b) summer-harvested soybean leaf.
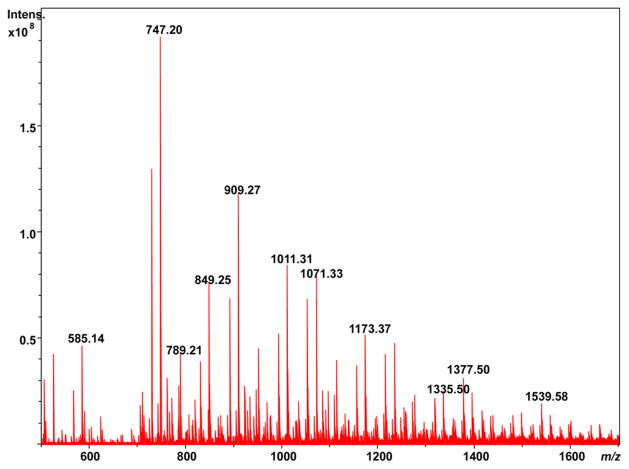
Since LDI does not use a matrix as MALDI does, there is potential for LDI to have higher lateral or spatial resolution for imaging MS. Our SolariX system possesses a camera and an ability to specify the area for laser desorption to occur. LDI of a single soybean trichome, a leaf hair projecting from the leaf epidermis, from a summer-harvested soybean leaf generates the mass spectrum shown in Figure 7. While the LDI mass spectrum of the trichome shows most of the same m/z as are found in Figure 6b, a compilation of random laser shots across the leaf, a greater number of higher m/z peaks beyond m/z 1300 are observed. This at least suggests that it may be possible to differentiate some anatomic features of the leaf using LDI imaging MS, similar to our recent report of MALDI imaging MS of a soybean leaf using two-dimensional graphene as a matrix,43 and is a subject worthy of future investigation.
Figure 7.
Direct LDI-FTICR mass spectrum of a soybean trichome.
CONCLUSIONS
Extractions of soybean leaves harvested at two different time points, midsummer and harvest day, show clear differences in the metabolite profile as determined by ESI-FTICR mass spectrometry. Moreover, when changes in species abundance relative to an internal standard are evaluated, a marked decrease in chlorophyll-related metabolites from summer to autumn is notable, along with a significant rise in the level of disaccharides. However, solubility limitations prevent many soybean metabolites from being detected in extracts. For many of these, MALDI or LDI may be used to examine directly the raw plant leaf tissue. MALDI generates a higher overall signal intensity than LDI for direct leaf profiling and also benefits from greater distinguishing features between autumn- and summer-harvested leaves than does LDI. LDI could be employed at different coordinates on the leaf surface, as shown for direct analysis of a leaf trichome (hair). Of course, with MALDI there is the potential for interference between the matrixes used and many potential low mass metabolites, so high-resolution mass analyzers would be preferable for MALDI of plant leaves.
Supplementary Material
Acknowledgments
The project described was supported by Award Number S10RR029517 from the National Center For Research Resources. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Center For Research Resources or the National Institutes of Health. We would like to thank Roger and Beverly Wood for permitting harvest of soybean leaves from their farm.
Footnotes
Notes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.anal-chem.5b03340.
Positive and negative ion ESI-FTICR mass spectra from summer-harvested leaves using other extraction solvents (PDF)
Table of fold change in m/z between seasons (ethanolic extract by ESI) (XLSX)
Peak lists of MALDI of soybean leaves (XLSX)
Peak lists of LDI of soybean leaves (XLSX)
References
- 1.Fiehn O, Kopka J, Dörmann P, Altmann T, Trethewey RN, Willmitzer L. Nat Biotechnol. 2000;18:1157–1161. doi: 10.1038/81137. [DOI] [PubMed] [Google Scholar]
- 2.Kaushik KA, Vareed SK, Basu S, Putluri V, Putluri N, Panzitt K, Brennan CA, Chinnaiyan AM, Vergara IA, Erho N, Weigel NL, Mitsiades N, Shojaie A, Palapattu G, Michailidis G, Sreekumar A. J Proteome Res. 2014;13:1088–1100. doi: 10.1021/pr401106h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bain JR, Stevens RD, Wenner BR, Ilkayeva O, Muoio DM, Newgard CB. Diabetes. 2009;58:2429–2443. doi: 10.2337/db09-0580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jansson J, Willing B, Lucio M, Fekete A, Dicksved J, Halfvarson J, Tysk C, Schmitt-Kopplin P. PLoS One. 2009;4:e6386. doi: 10.1371/journal.pone.0006386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffin JL, Atherton H, Shockcor J, Atzori L. Nat Rev Cardiol. 2011;8:630–643. doi: 10.1038/nrcardio.2011.138. [DOI] [PubMed] [Google Scholar]
- 6.Stitt M, Fernie AR. Curr Opin Biotechnol. 2003;14:136–144. doi: 10.1016/s0958-1669(03)00023-5. [DOI] [PubMed] [Google Scholar]
- 7.Glinski M, Weckwerth W. Mass Spectrom Rev. 2006;25:173–214. doi: 10.1002/mas.20063. [DOI] [PubMed] [Google Scholar]
- 8.Wilson ID, Plumb R, Granger J, Major H, Williams R, Lenz EM. J Chromatogr B: Anal Technol Biomed Life Sci. 2005;817:67–76. doi: 10.1016/j.jchromb.2004.07.045. [DOI] [PubMed] [Google Scholar]
- 9.Solanky KS, Bailey NJC, Beckwith-Hall BM, Davis A, Bingham S, Holmes E, Nicholson JK, Cassidy A. Anal Biochem. 2003;323:197–204. doi: 10.1016/j.ab.2003.08.028. [DOI] [PubMed] [Google Scholar]
- 10.Johnson HE, Broadhurst D, Kell DB, Theodorou MK, Merry RJ, Griffith GW. Appl Environ Microbiol. 2004;70:1583–1592. doi: 10.1128/AEM.70.3.1583-1592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jarvis RM, Goodacre R. FEMS Microbiol Lett. 2004;232:127–132. doi: 10.1016/S0378-1097(04)00040-0. [DOI] [PubMed] [Google Scholar]
- 12.Roessner-Tunali U, Hegemann B, Lytovchenko A, Carrari F, Bruedigam C, Granot D, Fernie AR. Plant Physiol. 2003;133:84–99. doi: 10.1104/pp.103.023572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia A, Barbas C. Methods Mol Biol. 2011;708:191–204. doi: 10.1007/978-1-61737-985-7_11. [DOI] [PubMed] [Google Scholar]
- 14.Becker S, Kortz L, Helmschrodt C, Thiery J, Ceglarek U. J Chromatogr B: Anal Technol Biomed Life Sci. 2012;883–884:68–75. doi: 10.1016/j.jchromb.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 15.Soga T. Methods Mol Biol. 2007;358:129–37. doi: 10.1007/978-1-59745-244-1_8. [DOI] [PubMed] [Google Scholar]
- 16.Tolstikov VV, Lommen A, Nakanishi K, Tanaka N, Fiehn O. Anal Chem. 2003;75:6737–6740. doi: 10.1021/ac034716z. [DOI] [PubMed] [Google Scholar]
- 17.Garratt LC, Linforth R, Taylor A, Lowe KC, Power JB, Davey MR. Plant Biotechnol J. 2005;3:165–174. doi: 10.1111/j.1467-7652.2004.00110.x. [DOI] [PubMed] [Google Scholar]
- 18.El-Zohri MHA, Cabala R, Frank H. Anal Bioanal Chem. 2005;382:1871–1876. doi: 10.1007/s00216-005-3331-0. [DOI] [PubMed] [Google Scholar]
- 19.Keurentjes JJB, Fu J, Ric, de Vos CH, Lommen A, Hall RD, Bino RJ, van der Plas LHW, Jansen RC, Vreugdenhil D, Koornneef M. Nat Genet. 2006;38:842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- 20.Fenselau C. Anal Chem. 1997;69:661–665. doi: 10.1021/ac971831z. [DOI] [PubMed] [Google Scholar]
- 21.Kahn ML, Parra-Colmenares A, Ford CL, Kaser F, McCaskill D, Ketchum RE. Anal Biochem. 2002;307:219–225. doi: 10.1016/s0003-2697(02)00046-5. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Garden RW, Sweedler JV. Trends Biotechnol. 2000;18:151–160. doi: 10.1016/s0167-7799(00)01427-x. [DOI] [PubMed] [Google Scholar]
- 23.Allen J, Davey HM, Broadhurst D, Heald JK, Rowland JJ, Oliver SG, Kell DB. Nat Biotechnol. 2003;21:692–696. doi: 10.1038/nbt823. [DOI] [PubMed] [Google Scholar]
- 24.Welti R, Wang X, Williams TD. Anal Biochem. 2003;314:149–152. doi: 10.1016/s0003-2697(02)00623-1. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Li W, Li M, Welti R. Physiol Plant. 2006;126:90–96. [Google Scholar]
- 26.Catharino RR, Haddad R, Cabrini LG, Cunha IBS, Sawaya ACHF, Eberlin MN. Anal Chem. 2005;77:7429–7433. doi: 10.1021/ac0512507. [DOI] [PubMed] [Google Scholar]
- 27.Han J, Danell RM, Gumerov DR, Scarlett CO, Speir JP, Parker CE, Rusyn I, Zeisel S, Borchers CH. Metabolomics. 2008;4:128–140. doi: 10.1007/s11306-008-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marshall AG. Int J Mass Spectrom. 2000;200:331–356. [Google Scholar]
- 29.Sung M, Kendall CWC, Koo MM, Rao AV. Nutr Cancer. 1995;23:259–270. doi: 10.1080/01635589509514380. [DOI] [PubMed] [Google Scholar]
- 30.Yoshiki Y, Kahara T, Okubo K, Sakabe T, Yamasaki T. Biosci, Biotechnol, Biochem. 2001;65:2162–2165. doi: 10.1271/bbb.65.2162. [DOI] [PubMed] [Google Scholar]
- 31.Fournier DB, Erdman JW, Jr, Gordon GB. Cancer Epidemiol Biomarkers PreV. 1998;7:1055–1065. [PubMed] [Google Scholar]
- 32.Xia J, Sinelnikov I, Han B, Wishart DS. Nucleic Acids Res. 2015;43:W251. doi: 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bricker DL, Russell DH. J Am Chem Soc. 1986;108:6174–6179. [Google Scholar]
- 34.Grese RP, Cerny RL, Gross ML, Senge M. J Am Soc Mass Spectrom. 1990;1:72–84. doi: 10.1016/1044-0305(90)80008-B. [DOI] [PubMed] [Google Scholar]
- 35.vanBreemen RB, Canjura FL, Schwartz SJ. J Agric Food Chem. 1991;39:1452–1456. [Google Scholar]
- 36.Milenkovic SM, Zvezdanovic JB, Andelkovic TD, Markovic DZ. Adv Technol. 2012;1:16–24. [Google Scholar]
- 37.Codex Alimentarius Commission. Draft Standard for Fat Spreads and Blended Spreads—Request for Comments on Food Additive Provisions. 2004 Jan; CL 2004/1-FO. [Google Scholar]
- 38.Xia YQ, Patel S, Bakhtiar R, Franklin RB, Doss GA. J Am Soc Mass Spectrom. 2005;16:417–421. doi: 10.1016/j.jasms.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 39.Quinn KD, Nguyen NQT, Wach MM, Wood TD. Rapid Commun Mass Spectrom. 2012;26:1767–1775. doi: 10.1002/rcm.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly M-A, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guao N, Zhang Y, Duggan GE, MacInnis GD, Weljie Al M, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L. Nucleic Acids Res. 2007;35:D521–526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karas M, Bachmann D, Hillenkamp F. Anal Chem. 1985;57:2935–2939. [Google Scholar]
- 42.Hölscher D, Shroff R, Knop K, Gottschaldt M, Crecelius A, Schneider B, Heckel DG, Schubert US, Svatoš A. Plant J. 2009;60:907–918. doi: 10.1111/j.1365-313X.2009.04012.x. [DOI] [PubMed] [Google Scholar]
- 43.Friesen WL, Schultz BJ, Destino JF, Alivio TEG, Steet JR, Banerjee S, Wood TD. J Am Soc Mass Spectrom. 2015;26:1963–1966. doi: 10.1007/s13361-015-1243-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.