Summary
Objectives
Discharging patients from the Neonatal Intensive Care Unit (NICU) can be delayed for non-medical reasons including the procurement of home medical equipment, parental education, and the need for children’s services. We previously created a model to identify patients that will be medically ready for discharge in the subsequent 2–10 days. In this study we use Natural Language Processing to improve upon that model and discern why the model performed poorly on certain patients.
Methods
We retrospectively examined the text of the Assessment and Plan section from daily progress notes of 4,693 patients (103,206 patient-days) from the NICU of a large, academic children’s hospital. A matrix was constructed using words from NICU notes (single words and bigrams) to train a supervised machine learning algorithm to determine the most important words differentiating poorly performing patients compared to well performing patients in our original discharge prediction model.
Results
NLP using a bag of words (BOW) analysis revealed several cohorts that performed poorly in our original model. These included patients with surgical diagnoses, pulmonary hypertension, retinopathy of prematurity, and psychosocial issues.
Discussion
The BOW approach aided in cohort discovery and will allow further refinement of our original discharge model prediction. Adequately identifying patients discharged home on g-tube feeds alone could improve the AUC of our original model by 0.02. Additionally, this approach identified social issues as a major cause for delayed discharge.
Conclusion
A BOW analysis provides a method to improve and refine our NICU discharge prediction model and could potentially avoid over 900 (0.9%) hospital days.
Keywords: Neonatal intensive care units, area under curve, patient discharge, ROC curve
1. Background and Objectives
Approximately four million babies are born in the United States each year and approximately 11% of those are born prematurely [1]. The cost of caring for these infants is substantial, with an estimated total annual cost of 26 billion dollars posing a significant financial burden for society in general and the health care system specifically [1]. Discharging these patients as soon as they are medically ready is critical for controlling expenditures.
Delayed discharge of hospitalized patients, who are medically ready for discharge, is a common occurrence and often related to dependency and the need for post-discharge services [2]. Neonates discharged from the NICU – whether they are premature or recovering from another condition – are prime examples of patients with dependencies on parents and caregivers, who rely heavily on post-discharge services for medical follow-up, home medical equipment, and home nursing [3]. Parents of these fragile infants require a significant amount of training and education regarding the special needs of their newborn, the use of medical equipment, and medication administration. Infants often require a number of services at the end of their hospitalization that may delay discharge including hearing screens, repeat state screens, immunizations, car seat testing, and eye exams. Finally, infants at risk for abuse and neglect, for example those with intra-uterine drug exposure, require clearance from Child Protective Services to ensure they are being discharged to a safe home environment.
While substantial work has been conducted predicting survival and outcomes in NICU patients[4, 5], we do not yet understand well how to predict discharge dates. Prior work suggests that discharge prediction is difficult using only perinatal factors, however may improve with additional clinical context, such as knowledge of later events or morbidities [6].
We previously described a predictive model using a Random Forest to analyze 26 clinical features extracted from the NICU attending physician daily progress note [3]. The goal of that model was to identify patients who would be medically ready for discharge in the next 10, 7, 4, and 2 days with the intent to make clinical staff aware and ready to address in advance the non-medical factors that often delay discharge of patients medically ready to go home.
The previous model performed well, achieving area under the curve (AUC) for the receiver operating characteristic (ROC) curve of 0.723, 0.754, 0.795, and 0.854 at 10, 7, 4 and 2 days until discharge, respectively. The model used structured and semi-structured data extracted from the attending physician progress note and it ignored the free text contained within the progress note. The goal of the work presented here is to use Natural Language Processing (NLP) to identify themes in our original model among those patients with missed discharge prediction and those who may have had a delayed discharge. These results could allow us to detect useful features missing from the original model that would allow for a more accurate model to classify NICU patients who are nearing discharge.
1.1 Related Work
NLP is frequently used to analyze medical documentation in order to identify patient cohorts. Yang et al. describes a text mining approach for obesity detection and later expanded it to extract medication information [7, 8]. Jiang et al. examined different machine learning algorithms to identify clinical entities from discharge summaries.[9] Wright et al. used an NLP support vector machine to categorize free text notes in order to identify patients with diabetes [10]. In 2012, Cui et al. used discharge summaries to effectively extract information regarding epilepsy and seizures [11]. Cosmin et al. describe an NLP system to identify ICU patients, who were diagnosed with pneumonia at any point in their hospital stay [12].
In pediatric settings, NLP has been used to identify asthma status[13], celiac disease[14], and epilepsy [15]. These studies demonstrate that NLP can be used to accurately identify patients belonging to certain cohorts; however, to our knowledge, we are presenting the first study using NLP in predicting discharge dates in an NICU.
Typically, when using NLP to evaluate the accuracy of a model, the results are compared to a known set of similar documents. This allows for the evaluation of precision (analogous to positive predictive value), recall (analogous to sensitivity), and F-score (a combined measure that assesses the tradeoff between precision and recall). We propose to use NLP for cohort discovery. It is our hypothesis that NLP can assist us in refining our NICU prediction model and identify patient characteristics from the clinical note that may be missing in our original NICU discharge prediction model.
2. Methods
2.1 Patients and Setting
We conducted a retrospective study of 6,302 patients admitted to the NICU at a large academic medical center from June 2007 to May 2013.
2.2 Exclusion Criteria
All patients admitted to the NICU were considered for the study. Since this project was part of a larger study, the exclusion criteria were the same as the original study: Patients who were back-transferred to another facility or who died during the course of their NICU hospitalization were excluded from the analysis. Also excluded from the analysis were patients with any missing daily neonatology progress notes.
2.3 Data Collection and Extraction
A large database containing all daily progress notes written by neonatology attending physicians was made available to the investigators [16]. The data from the progress notes were in a semi-structured text format and were extracted using regular expressions in Python (version 2.7.3) and SQL. In addition, these data were cross-referenced with the enterprise data warehouse in order to obtain basic patient information such as date of birth and ICD-9 codes used for billing during the hospitalization.
2.4 Feature Descriptions
Our original predictive model included the clinical features listed in ► Table 1 [3].
Table 1.
Features used in the Predictive Model
| Quantitative Features (Unit of Measure) | Qualitative Features (Unit of Measure) | Engineered Features (Unit of Measure) | Sub-Population Features |
|---|---|---|---|
| Weight (kg) | On Infused Medication (Y/N) | Number of Days Since Last A&B Event (days) | Premature (Y/N) |
| Birth Weight (kg) | On Caffeine (Y/N) | Number of Days Off Infused Medication (days) | Cardiac Surgery (Y/N) |
| Apnea and Bradycardia (A&B) Events (number) | On Ventilator (Y/N) | Number of Days Off Caffeine (days) | GI Surgery (Y/N) |
| Amount of Oral Feeds (ml) | Number of Days Off Ventilator (days) | Neurosurgery (Y/N) | |
| Amount of Tube Feeds (ml) | Number of Days Off Oxygen (days) | ||
| Percentage of Oral Feeds (%) | Number of Days Percent of Oral Feeds >90% (days) | ||
| Gestational Age (weeks) | Total Feeds (Oral + Tube Feeds) (ml) | ||
| Gestational Age at Birth (weeks) | Ratio of Weight to Birth Weight | ||
| Day of Life (days) | Amount of Oral Feeds/Weight (ml/kg/day) | ||
| Oxygen (per liter) |
All of the clinical features listed in ► Table 1 were extracted using structured or semi-structured sections of the progress note – not the Assessment and Plan. For the NLP evaluation, we used only the Assessment and Plan section of the daily progress note. This section tends to contain the most relevant clinical information conveying the provider’s understanding of the patient’s problems and her treatment plans.
The entire text of the Assessment and Plan section was extracted and tokenized using Python’s natural language toolkit (version 3.0.1) [17]. All of the stop words and numbers were removed. Additionally, words were converted to all lower case and only words with a length greater than or equal to three characters were considered in the corpus. This provided a simple “bag of words” (single words and bigrams). Negation was not considered in this approach.
2.5 Original Model – Matrix Organization
Each row in a matrix represents one hospital day for a patient (i.e., a patient-day). Therefore, if the patient was in the hospital for 20 days, that patient occupied 20 rows of the matrix.
The original model’s matrix contained 26 columns and N rows, one column for each extracted feature from ► Table 1 and one row for each patient-day. Some columns contained boolean values, while other represented real values.
2.6 Original Model – Discharge Prediction
The objective of the original work was to train a classifier to identify the days to discharge. In the original model, a boolean dependent vector was constructed of length N (one index for each patient-day) such that index i was ‘1’ when i was equal to the desired days to discharge, and ‘0’ otherwise. If there were K patients in our population, there would be K indexes in the vector with a value of ‘1’.
In the initial work, we used the Original Model’s matrix and Discharge Prediction dependent vector to train a random forest classifier to predict days to discharge at 2, 4, 7 and 10 days. For each patient-day, the classifier outputs a probability for discharge.
Expanding on our previous work, this paper takes these prediction probabilities and uses them to identify patients that performed poorly under the original model. We defined poorly performing patients in two ways using Days to Discharge (DTD) = 4: (i) Missed Discharge Predictions: patient days with a probability of 0.2 or less for that patient being discharged in the next two days, and (ii) Delayed Discharges: patient days with a probability of 0.5 or greater at days to discharge of 10 or more. ► Figure 1 provides an overview of this analysis.
Fig. 1.
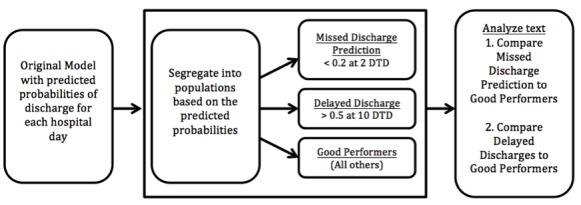
Workflow diagram demonstrating process for cohort discovery. DTD = Days to Discharge.
2.7 Bag-of-Words Model Matrix
Next we examined if note text could predict NICU discharge. To this end, a BOW matrix was constructed with 560 columns and N rows, because there were 560 unique words in the data set and N patient-days. A column represented a single word. If the word appeared in the Assessment and Plan section of the progress note on the day represented by that particular row, a ‘1’ was assigned to the field representing the progress note (repeated words were not given more weight than a single occurrence of a word). If the word was not present, a ‘0’ was assigned.
2.8 Model Comparison
We ran tests to analyze which model (the Original or BOW) was best suited to train a classifier with the Discharge Prediction dependent vector to predict the days to discharge at 2, 4, 7, and 10 days.
2.9 Interpreting Poor Prediction Results
Given the set of poorly performing patients, we wanted to analyze the Assessment and Plan text to identify the reasons for classification errors. To this end, we used the BOW matrix, but instead constructed two new dependent vectors for Cohort Discovery: (i) When building the matrix for Missed Discharge Prediction patients, a Missed Discharge Prediction dependent vector set all Missed Discharge Prediction patients’ rows to ‘1’, and all other rows to ‘0’. Therefore, if a Missed Discharge Prediction patient was in the NICU for 20 days, there would be 20 1’s for the patient in the vector. (ii) Similarly, we constructed a Delayed Discharge Prediction dependent vector that set all Delayed Discharge patients’ rows to ‘1’, and all other rows to ‘0’. We then trained a classifier for each poorly performing cohort using this new dependent vector. This allowed us to identify the most important words that differentiated the two poorly performing patients in the Original Model with other, well-performing patients. ► Figure 2 provides an overview of the various matrices.
Fig. 2.
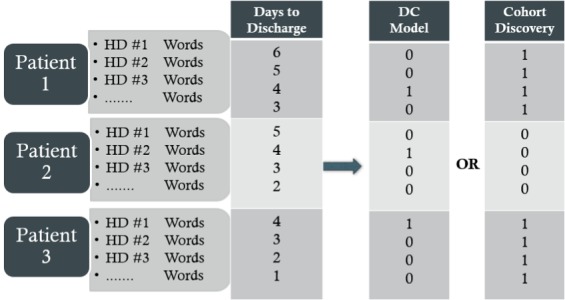
Construction of matrix and model vector for prediciting days to discharge or cohort discovery. HD = Hospital Day.
2.10 Data Analysis
A supervised machine learning approach using a Random Forest Classifier (RF) in Python’s Sci-kit Learn module (version 0.15.2) [18] was used to analyze the data and build a model. A RF constructs many binary decision trees that branch based on randomly chosen features. The RF in Sci-kit Learn uses an optimized Classification And Regression Trees (CART) algorithm for constructing binary trees using the features and thresholds (values) that yield the largest information gain at each node. The Sci-kit Learn package allows for the selection of either the gini impurity or entropy algorithms to determine feature importance. These algorithms performed similarly and we chose to use gini impurity because it attempts to minimize the probability of misclassification (as opposed to the impurity of a split) and, therefore, is slightly more robust to misclassifications. We used the same Random Forest approach in our original model.
Models were trained using different combinations of DTD (2, 4, 7, 10 days) and different populations of poorly performing patients. Using our original prediction model, we were able to determine poorly performing patients by evaluating their predicted probability of discharge. For example, we ran our initial model predicting which patients were within 4 days of discharge from the NICU. We obtained the predicted probability (ranging from 0 to 1) that our model assigned to each patient for each hospital day. If our model assigned a probability of 0.2 or less of discharge when the patient was actually 2 days from discharge, we then would consider this a poorly performing patient in the category of Missed Discharge Prediction. Additionally, if our model assigned a probability of 0.5 or higher when the patient was 10 days or more from discharge, these poorly performing patients were considered Delayed Discharge patients (► Figures 3–5).
Fig. 3.
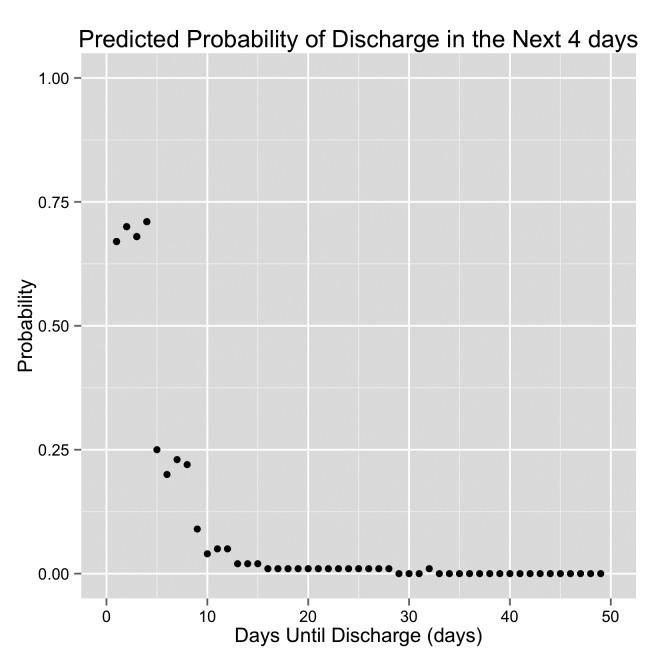
Graph demonstrating the predicted probability of discharge by the original model. DTD is displayed on the x-axis. The patient is discharged when DTD = 0 (the left side of each graph). The right side of each graph are days early in the hospital stay. This is an example of a “Good Performing” patient.
Fig. 4.
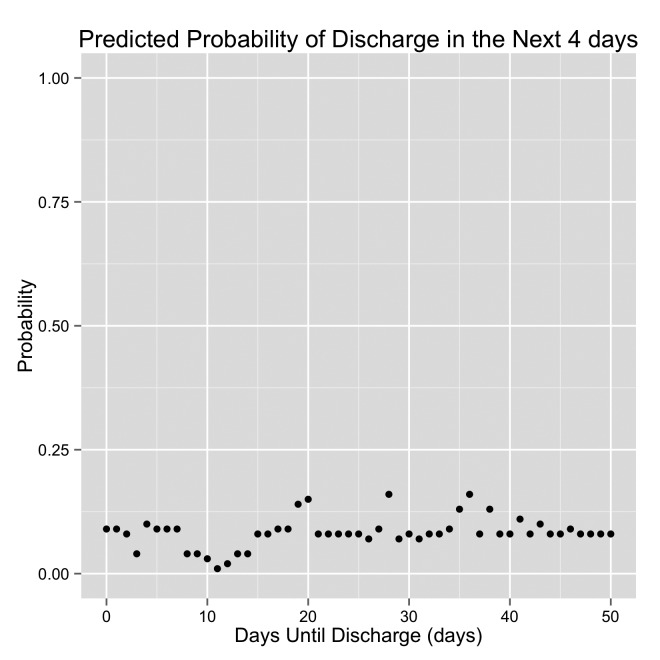
Graph demonstrating the predicted probability of discharge by the original model. DTD is displayed on the x-axis. The patient is discharged when DTD = 0 (the left side of each graph). The right side of each graph are days early in the hospital stay. This is an example of a “Missed Discharge” patient.
Fig. 5.
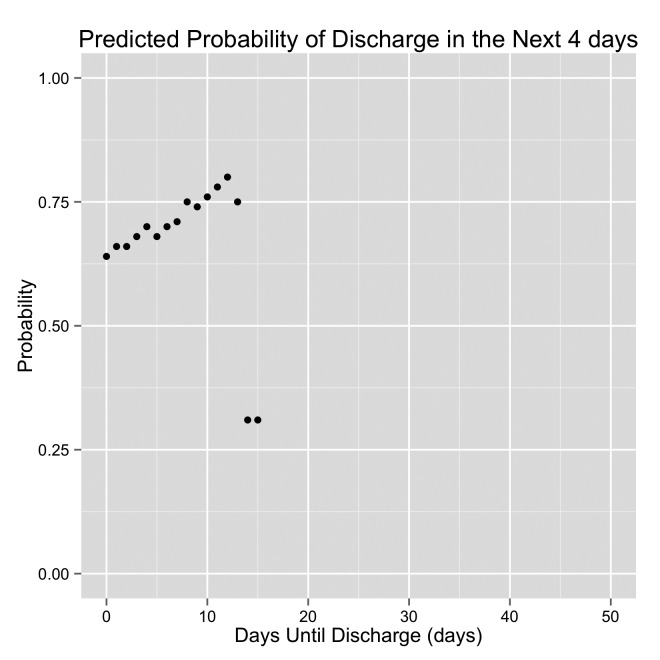
Graph demonstrating the predicted probability of discharge by the original model. DTD is displayed on the x-axis. The patient is discharged when DTD = 0 (the left side of each graph). The right side of each graph are days early in the hospital stay. This is an example of a “Delayed Discharge” patient.
2.11 Cross Validation
Each time a model was run, half of the patients (and all their associated daily rows) were randomized into a training set and the remaining patients were assigned to the testing set. Since the number of poorly performing patients in the sample was relatively small, halving the data provided both testing and training sets an adequate number of patients of interest. To achieve small enough standard deviations, the patients were randomized a total of five times for each model and the AUC for the ROC curve was obtained for each testing set. The reported AUC is the average of the five AUC’s obtained after each round of randomization. Additionally, each time a model was run, the top 20 words used in the model were ranked in order of importance.
IRB Approval
The Institutional Review Board of Vanderbilt University approved this study.
3. Results
The initial database consisted of 6,302 patients admitted to the NICU between June 2007 and May 2013. There were 256 deaths during this time period. A total of 1,154 patients were excluded because the database did not contain physician progress notes for every day of their hospital course. There were 199 patients back-transferred to other NICU’s in the region. The final matrix consisted of 4,693 unique patients accounting for 103,206 hospital days with a mean LOS of 30 days.
3.1 Bag of Words for Discharge Prediction
► Table 2 shows the results of the original model only, bag of words (BOW) using only words from the Assessment and Plan, and the combined approach with regards to discharge classification.
Table 2.
Comparing discharge prediction models among the original model, BOW model and the combination of the two models. BOW = bag of words.
| Days Until Discharge (days) | Original Model (AUC) | BOW Model (AUC) | Combined Original and BOW (AUC) |
|---|---|---|---|
| 10 | 0.723 | 0.569 | 0.633 |
| 7 | 0.754 | 0.589 | 0.677 |
| 4 | 0.795 | 0.654 | 0.752 |
| 2 | 0.854 | 0.743 | 0.837 |
► Table 3 shows the top 15 most important bigrams for predicting discharge at 2, 4, 7, and 10 days until discharge.
Table 3.
The top 15 most important (listed in order) bigrams for each of the days to discharge
| Days Until Discharge (days) | Most important Bigrams |
|---|---|
| 10 | continue monitor, today continue, pcv retic, enteral feeds, day continue, total fluids, prior discharge, feeds day, weight gain, continue follow, past hrs, full feeds, updated bedside, wean today, room air |
| 7 | continue monitor, weight gain, prior discharge, today continue, pcv retic, full feeds, enteral feeds, feeds day, next week, day continue, past hours, amp gent, may need, continue follow, past hrs |
| 4 | prior discharge, continue monitor, weight gain, pcv retic, today continue, feeds day, past hrs, day continue, cbc crp, amp gent, room air, follow clinically, past hours, discharge home, continue follow |
| 2 | weight gain, prior discharge, continue monitor, full feeds, pcv retic, hearing screen, room air, amp gent, fen lib, repeat echo, cbc crp, continue follow, today continue, last hours, follow clinically. |
3.2 Bag of Words for Cohort Discovery – Missed Discharge Prediction patients with Probability less than 0.2 at 2 or less DTD
We extracted the most important words as determined by the bag of words model when comparing patients who performed well in our original model to those that performed poorly in the Missed Discharge Prediction cohort in our original model. There were 194 patients categorized as poor performers using these criteria.
► Table 4 shows the most significant words differentiating well performing from poorly performing patients with a probability of 0.2 or less to be discharged in the next two days. The words are listed in order of importance and a few words have been excluded because of inability to determine the context (for example, “continue monitor”, and “per protocol”).
Table 4.
The most important single words and bigram in order of importance differentiating poorly performing patients in the Missed Discharge cohort (probability of less than 0.2 at 2 or less days until discharge) from well performing patients in our original model.
| Single Words | Bigrams |
|---|---|
| fistula, ent, tube, esophageal, atresia, nissen, vfss, breech, psychosocial, uti, gtube, aspiration, hus, reflux, vcug | status post, esophageal atresia, repeat echo, pulmonary hypertension, enteral feeds, lung disease, goal sats, urine culture, infectious disease, drug screen, plus disease, stage zone, room air |
3.3 Bag of Words for Cohort Discovery – Delayed Discharge Patients with Probability more than 0.5 at 10 or more DTD
► Table 5 lists the most significant words differentiating poorly performing patients in the Delayed Discharge cohort with a probability of 0.5 or higher at 10 or more days until discharge. There were 190 patients categorized as poor performers using these criteria.
Table 5.
The most important single words and bigram in order of importance differentiating poorly performing patients in the Delayed Discharge cohort (probability of more than 0.5 at 10 or more days until discharge) from well performing patients in our original model.
| Single Words | Bigrams |
|---|---|
| hep, social, weight, daily, restarted, signs, direct, endocrine, positive, drug, mother, birth, dcs, congenital, syndrome, continue, prematurity | social work, work breathing, low birth, birth weight, initial cbc, clinical signs, room air, dcs involved, possible sepsis, prior discharge, infectious disease, monitor respiratory, continue monitor, hearing screen, newborn screen, meconium drug, drug screen |
4. Discussion
4.1 Bag of Words for Discharge Prediction
The bag of words approach performed poorly with regards to discharge prediction. This may be explained by the fact that only a very small part of the progress note (the Assessment and Plan section) was used as the corpus. Second, because our original model contained quantitative clinical data, we excluded any numerical values from our NLP analysis.
4.2 Bag of Words for Missed Discharge Prediction Cohort Discovery – Probability less than 0.2 at 2 or less DTD
Using a bag of words model for cohort discovery identified characteristics for some patients that are not performing well in our original model (► Table 4).
First, our original model is not performing well on some surgical patients. The top two most important bigrams are “status post” and “esophageal atresia”. Additionally, four of the most important single words are “fistula”, “esophageal”, “atresia”, and “Nissen”. All of these words would be found in patients who have a gastrointestinal abnormality requiring surgery or have had a surgical repair already performed. Feeding difficulties and subsequent increased length of stay have been described in this population [19]. Also, patients who have had a “Nissen” procedure likely needed the procedure because of reflux with aspiration pneumonia. The words “aspiration”, “reflux”, “gtube” and “vfss” (swallow study) are likely related to this GI surgery. Finally, one of the most important single words is “ENT”. Neonates can have congenital anomalies of their ear, nose, or throat requiring surgical correction; therefore, capturing these patients in our model could help improve it.
Another interesting combination of words for cohort discovery is “psychosocial” and “drug screen”. The importance of these words suggests that our model is not performing well on patients, who may have had intrauterine drug exposure or whose parents may have had psychosocial issues.
Our model also appears to perform poorly on patients who have a history of “pulmonary hypertension”. These patients tend to be very sick early in their hospital stay and may require extra-corporeal membrane oxygenation (ECMO). While these patients have significantly improved clinical status when they are two days from discharge, it appears that our model is not correctly capturing the improved clinical status of these patients.
Finally, the two bigrams “plus disease” and “stage zone” are references to retinopathy of prematurity – which has been associated with severity of prematurity [20, 21]. Premature infants with retinopathy of prematurity (ROP) need to have an eye exam performed by an ophthalmologist near the time of their discharge. The presence of these words in the Assessment and Plan could be referencing the results of this last exam before discharge or the need to schedule an examination prior to discharge.
4.3 Bag of Words for Delayed Discharge Cohort Discovery – Probability more than 0.5 at 10 or more DTD
Using a bag of words approach on these patients helped identify possible reasons for delayed discharges (► Table 5). First, social factors appear to be an issue. Words such as “social”, “drug”, and “dcs” (Department of Children’s Services) indicate social and/or custody issues may be causing discharge delays in patients who are medically ready for discharge. This is further supported by the bigrams “social work”, “dcs involved”, “meconium drug”, and “drug screen”.
In addition to our original model predicting a greater than 0.5 probability of discharge for these patients, the bag of words also supports their readiness for discharge. Words from ► Table 3 (important words for discharge prediction) such as “prior discharge”, “continue monitor”, “room air”, “hearing screen” also appear in ► Table 5 – the list of important words for patients who may be ready for discharge, but are delayed. In our data set, there were 904 hospital days (194 patients) that met these probability criteria.
4.4 Further Evaluation
The bag of words approach identified patient characteristics that were not present in our original model mainly pertaining to specific diagnoses that lead to feeding problems or need for prolonged monitoring like ROP. Using this knowledge in our model, we will be able to add other features that will aid in capturing and improving the predictive accuracy for poorly performing patients. For example, our model could identify patients that have had a social work consult performed. We could also use ICD-9 codes to capture patients who have esophageal atresia, pulmonary hypertension, or retinopathy of prematurity.
In our original model, important predictive factors centered around feeding – in particular oral feeding. If the infant was consistently consuming a large part of her/his feeds orally, then she/he was nearing discharge. This NLP analysis would indicate that our model is not performing well on patients who go home on g-tube feedings. Therefore, we performed the following test to determine the impact on our model if we correctly classified those patients being discharged on g-tube feeds:
We used the NLP bag of words approach and identified all patients who had the words “gtube” or “g-tube” in Assessment and Plan of their progress note.
We set the dependent prediction vector in our original model to “1” instead of “0” for the “g-tube” patients
We ran our original model as normal and measured the change in prediction accuracy.
The result of this manipulation of the output vector is shown in ► Table 6.
Table 6.
Original model improved with NLP for “G-tube” patients. Improvements suggest that we were able to correctly capture and classify all patients discharged on g-tube feeds.
| Days Until Discharge (days) | Original Model (AUC) | Correctly classified g-tube patients | |
|---|---|---|---|
| (AUC) | (difference) | ||
| 10 | 0.723 | 0.741 | (+ 0.018) |
| 7 | 0.754 | 0.775 | (+ 0.021) |
| 4 | 0.795 | 0.817 | (+ 0.022) |
| 2 | 0.854 | 0.863 | (+ 0.009) |
► Table 6 demonstrates that correctly classifying patients, who are discharged home on g-tube feeds, improves the accuracy of our original predictive model.
4.5 Limitations and Next Steps
One limitation of this study is that we only used the Assessment and Plan section of the attending physician progress note in the bag of words model. It is likely that more information from the use of the entire progress note would benefit the accuracy of our predictive model.
Another limitation is that even though NLP identified cohorts that do not perform well in our original model, it may be difficult to find ways to integrate those cohorts in our original model. For example, some patients who are discharged home on g-tube feeds may actually look different clinically. Some patients may be able to take a portion of their feedings orally while others will be reliant on continuous g-tube feedings.
A third limitation with the NLP analysis performed is that not all patients may be correctly classified. For example, while we identified a significant word as “vfss”, there may be other patients in whom “swallow study” is actually written out in the assessment and plan. Capturing all the ways in which medical professionals abbreviate is a difficult task and can cause some patients to be misclassified. The lack of standard conventions in clinical text has been identified as a barrier to NLP analysis [22].
A fourth limitation is that we trained the model using actual discharge dates. Some of these patients may have been ready for discharge sooner and we may have improved model performance if we could identify and adjust for these patients. Additionally, our model may predict discharge too early, which could lead to re-admissions or unmet parental expectations.
Omitting negations in our analysis is another limitation. A patient may have been described as “no reflux” and might have been lumped in our analysis as a reflux patient.
This study depends and builds upon our prior work. While this type of NLP analysis may not be generalizable to every institution, almost all of the data from our original model should be available in most NICU’s and could be utilized in those hospitals. The generalizability to other units and patient populations may be limited by the unique clinical characteristics of our NICU population.
While this study was a simple application of NLP and did not improve discharge prediction, the next steps in the refinement of our NICU discharge prediction model will be to use these cohorts discovered through our bag of words analysis and modify our original prediction model to include features related to these cohorts. For example, we could use ICD-9 codes to capture patients with pulmonary hypertension and retinopathy of prematurity to determine if there are other features that can be used to more accurately classify these patients.
5. Conclusions
An NLP analysis using a simple bag of words approach can be effectively used to discover under-performing cohorts and delayed discharges in a NICU discharge prediction model. Correctly classifying these cohorts can then be used to improve the predictive accuracy of the model.
Acknowledgments
The authors appreciate the assistance of the Research Derivative Team for assisting with the retrieval of data.
The publication described was supported by CTSA award No. UL1TR000445 from the National Center for Advancing Translational Sciences. Its contents are solely the responsibility of the authors and do not necessarily represent official views of the National Center for Advancing Translational Sciences or the National Institutes of Health.
Footnotes
Clinical Relevance
The results of this study inform clinicians regarding NICU patient populations that may have delayed discharge when they are medically ready for discharge. Identifying these populations will raise awareness for clinicians and, hopefully, prevent discharge delay in these infants.
Abbreviations
AUC – Area under the Curve, CART -- Classification And Regression Trees, DTD – Days to Discharge, GI – Gastrointestinal, LOS – Length of Stay, NICU – Neonatal Intensive Care Unit, NS – Neurosurgery, RF – Random Forest.
Funding Source
National Library of Medicine Training Grant 5T15LM007450–13.
Financial Disclosure
Dr. Lehman serves in a part-time role at the American Academy of Pediatrics. He also received royalties for the textbook Pediatric Informatics, and travel funds from the American Medical Informatics Association, the International Medical Informatics Association and the World Congress on Information Technology.
Dr. Fabbri has an equity interest in Maize Analytics, LLC. Dr. Temple has no financial disclosures.
Conflict of Interest
The authors have no conflicts of interest to disclose.
Protection of Human and Animal Subjects
The study was performed in compliance with the World Medical Association Declaration of Helsinki on Ethical Principles for Medical Research Involving Human Subjects, and was reviewed by the Vanderbilt University Institutional Review Board.
References
- 1.Bockli K, Andrews B, Pellerite M, Meadow W. Trends and challenges in United States neonatal intensive care units follow-up clinics. Journal of perinatology : official journal of the California Perinatal Association 2014; 34(1): 71-74. [DOI] [PubMed] [Google Scholar]
- 2.Challis D, Hughes J, Xie C, Jolley D. An examination of factors influencing delayed discharge of older people from hospital. International journal of geriatric psychiatry 2014; 29(2): 160-168. [DOI] [PubMed] [Google Scholar]
- 3.Temple MW, Lehmann CU, Fabbri D. Predicting Discharge Dates From the NICU Using Progress Note Data. Pediatrics 2015; 136(2):e395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manktelow BN, Seaton SE, Field DJ, Draper ES. Population-based estimates of in-unit survival for very preterm infants. Pediatrics 2013; 131(2): e425-e432. [DOI] [PubMed] [Google Scholar]
- 5.Draper ES, Manktelow B, Field DJ, James D. Prediction of survival for preterm births by weight and gestational age: retrospective population based study. Bmj 1999; 319(7217): 1093-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hintz SR, Bann CM, Ambalavanan N, Cotten CM, Das A, Higgins RD, et al. Predicting time to hospital discharge for extremely preterm infants. Pediatrics 2010; 125(1): e146-e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang H, Spasic I, Keane JA, Nenadic G. A text mining approach to the prediction of disease status from clinical discharge summaries. Journal of the American Medical Informatics Association: JAMIA 2009; 16(4):596–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H. Automatic extraction of medication information from medical discharge summaries. Journal of the American Medical Informatics Association: JAMIA 2010; 17(5): 545-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang M, Chen Y, Liu M, Rosenbloom ST, Mani S, Denny JC, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. Journal of the American Medical Informatics Association: JAMIA 2011; 18(5): 601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright A, McCoy AB, Henkin S, Kale A, Sittig DF. Use of a support vector machine for categorizing free-text notes: assessment of accuracy across two institutions. Journal of the American Medical Informatics Association: JAMIA 2013; 20(5): 887-890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui L, Bozorgi A, Lhatoo SD, Zhang GQ, Sahoo SS. EpiDEA: extracting structured epilepsy and seizure information from patient discharge summaries for cohort identification. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium 2012; 2012: 1191-1200. [PMC free article] [PubMed] [Google Scholar]
- 12.Bejan CA, Vanderwende L, Evans HL, Wurfel MM, Yetisgen-Yildiz M. On-time clinical phenotype prediction based on narrative reports. AMIA Annual Symposium proceedings/AMIA Symposium AMIA Symposium 2013; 2013: 103-110. [PMC free article] [PubMed] [Google Scholar]
- 13.Wu ST, Juhn YJ, Sohn S, Liu H. Patient-level temporal aggregation for text-based asthma status ascertainment. Journal of the American Medical Informatics Association: JAMIA 2014; 21(5): 876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludvigsson JF, Pathak J, Murphy S, Durski M, Kirsch PS, Chute CG, et al. Use of computerized algorithm to identify individuals in need of testing for celiac disease. Journal of the American Medical Informatics Association: JAMIA 2013; 20(e2): e306-e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Connolly B, Matykiewicz P, Bretonnel Cohen K, Standridge SM, Glauser TA, Dlugos DJ, et al. Assessing the similarity of surface linguistic features related to epilepsy across pediatric hospitals. Journal of the American Medical Informatics Association: JAMIA 2014; 21(5): 866-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Danciu I, Cowan JD, Basford M, Wang X, Saip A, Osgood S, et al. Secondary use of clinical data: The Vanderbilt approach. Journal of biomedical informatics 2014; 52(0):28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. http://www.nltk.org.
- 18. http://scikit-learn.org/stable/index.html.
- 19.Wang J, Du L, Cai W, Pan W, Yan W. Prolonged feeding difficulties after surgical correction of intestinal atresia: a 13-year experience. Journal of pediatric surgery 2014; 49(11): 1593-1597. [DOI] [PubMed] [Google Scholar]
- 20.Garg R, Agthe AG, Donohue PK, Lehmann CU. Hyperglycemia and retinopathy of prematurity in very low birth weight infants. Journal of perinatology: official journal of the California Perinatal Association 2003; 23(3): 186-194. [DOI] [PubMed] [Google Scholar]
- 21.Chavez-Valdez R, McGowan J, Cannon E, Lehmann CU. Contribution of early glycemic status in the development of severe retinopathy of prematurity in a cohort of ELBW infants. Journal of perinatology: official journal of the California Perinatal Association 2011; 31(12): 749-756. [DOI] [PubMed] [Google Scholar]
- 22.Chapman WW, Nadkarni PM, Hirschman L, D’Avolio LW, Savova GK, Uzuner O. Overcoming barriers to NLP for clinical text: the role of shared tasks and the need for additional creative solutions 2011; 2011–09–01 00:00:00. 540–3 p. [DOI] [PMC free article] [PubMed] [Google Scholar]