Fig. 11.
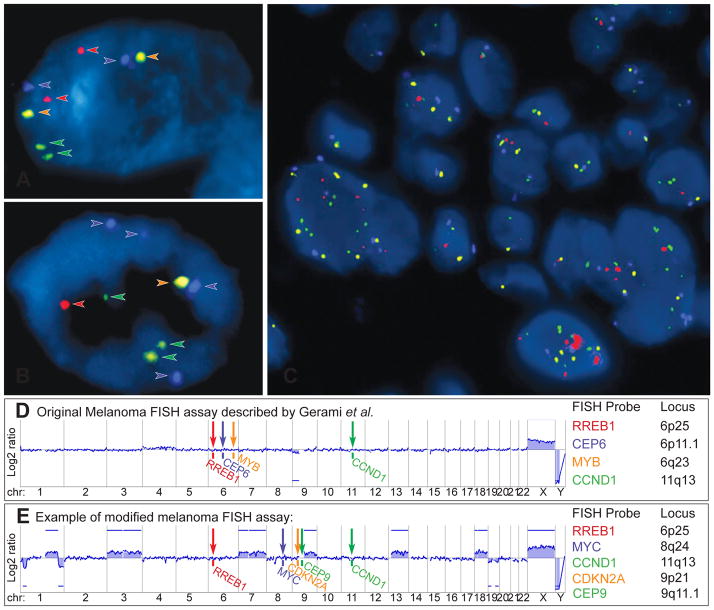
Fluorescence in situ hybridisation (FISH) as ancillary diagnostic tool for ambiguous melanocytic tumours. Melanoma FISH assays are often based on 4 probes, which are labelled with 4 different fluorescent dyes (red, green, orange, aqua) and bind to 4 specific chromosomal regions. (A) In normal cells without chromosomal gains and losses 2 signals of each probe are detected. (B) In cells with chromosomal aberrations, which affect the FISH probe-binding regions, losses or gains of the FISH signals can be observed (here 4 aqua, 3 green, and 1 red and orange signal). (C) FISH of a melanoma showing several cell nuclei with a varying number of FISH probes indicating chromosomal aberrations and chromosomal instability. (D) Binding sites of the original 4 FISH probes described by Gerami et al.: RREB1 (6p25), MYB (6q23), CCND1 (11q13) genes and of centromere 6. This aCGH profile of an AST shows loss of chromosome arm 9p, which would not be detected by FISH. (E) Modified and commercially used assay with FISH probes for RREB1 (6p25), MYC (8q24), CDKN2A (9p21), centromere 9 (CEP 9) and CCND1 (11q13). The corresponding aCGH profile detects several chromosomal aberrations involving chromosome 1, 3, 7, 9, 13, 18, 19, all of which would be missed by FISH. These examples illustrate that melanoma FISH has major blind spots. On the other hand, the detection of a chromosomal aberration by FISH does not prove that the tumour is malignant (see Fig. 9). Consequently, FISH is not very helpful in the diagnosis of ASTs.