Abstract
This review (with 90 refs.) covers the state of the art in optofluidic devices with integrated solid-state nanopores for use in detection and sensing. Following an introduction into principles of optofluidics and solid-state nanopore technology, we discuss features of solid-state nanopore based assays using optofluidics. This includes the incorporation of solid-state nanopores into optofluidic platforms based on liquid-core anti-resonant reflecting optical waveguides (ARROWs), methods for their fabrication, aspects of single particle detection and particle manipulation. We then describe the new functionalities provided by solid-state nanopores integrated into optofluidic chips, in particular acting as smart gates for correlated electro-optical detection and discrimination of nanoparticles. This enables the identification of viruses and λ-DNA, particle trajectory simulations, enhancing sensitivity by tuning the shape of nanopores. The review concludes with a summary and an outlook.
Keywords: Single biomolecule detection, Particle manipulation, Liquid core waveguide, ARROW waveguides, Bioassay, System integration, Electro-optics, Fluorescence analysis
Introduction
Optofluidics
The miniaturization and automation of analytical tools is a very active area of research and development. The appearance of microfluidics has intensified this trend, thanks to microfluidic devices’ compactness and their ability to control fluid flow. [1–5] Building on the success of microfluidics, the motivation to append advanced functions and increasing device sensitivity using optical methods led to a newly emerging field, optofluidics. Optofluidics is the seamless integration of microfluidics, optical analysis and manipulation methods. Within a single optofluidic device, the flexibility and controllability of fluid flow can simplify the transportation of biological samples and modify light propagation by changing the speed, direction, and refractive index of fluids. At the same time, optical detection methods bring high sensitivity and excellent spatial resolution. Due to the capability of controlling fluid and light at the same time within a miniscule volume, we have seen optofluidic devices used in a broad range of applications such as on chip lasers, sensors, optofluidic microscopes, etc. [6–12] Especially in the field of bioanalysis, the rising interest in point-of-care devices has generated numerous reports of successful applications of optofluidics in biosensing and bioanalysis, which makes optofluidic devices a great candidate for substituting conventional bulky equipment.
Integration of functional structures into optofluidic devices
Optofluidic devices are also perfect platforms for integrating modern micro/nano-scale fluidic or optical structures, which will further boost the performance of optofluidics in the field of bioanalysis sciences. Microfluidic channels and valve networks can be used for precise control of liquid flow, which leads to improvements in upstream sample preparation. [13] At the same time, the integration of optical structures, such as plasmonic nanostructures, gratings, photonic crystal structures, resonators, etc., [14–19] greatly increases detection sensitivity and reduces the limit of detection for devices. However, the functional extension of optofluidics is not only restricted to microfluidic or optical structures. In recent years, the use of electrical techniques has drawn the attention of many researchers. [20–22] In addition to being used for manipulating particles or tuning optical properties, in this article we will show that electrical elements can also be used for sensitive detection when integrated into optofluidics, offering the potential for multi-modal analysis at the single particle level.
Solid-state nanopore technology
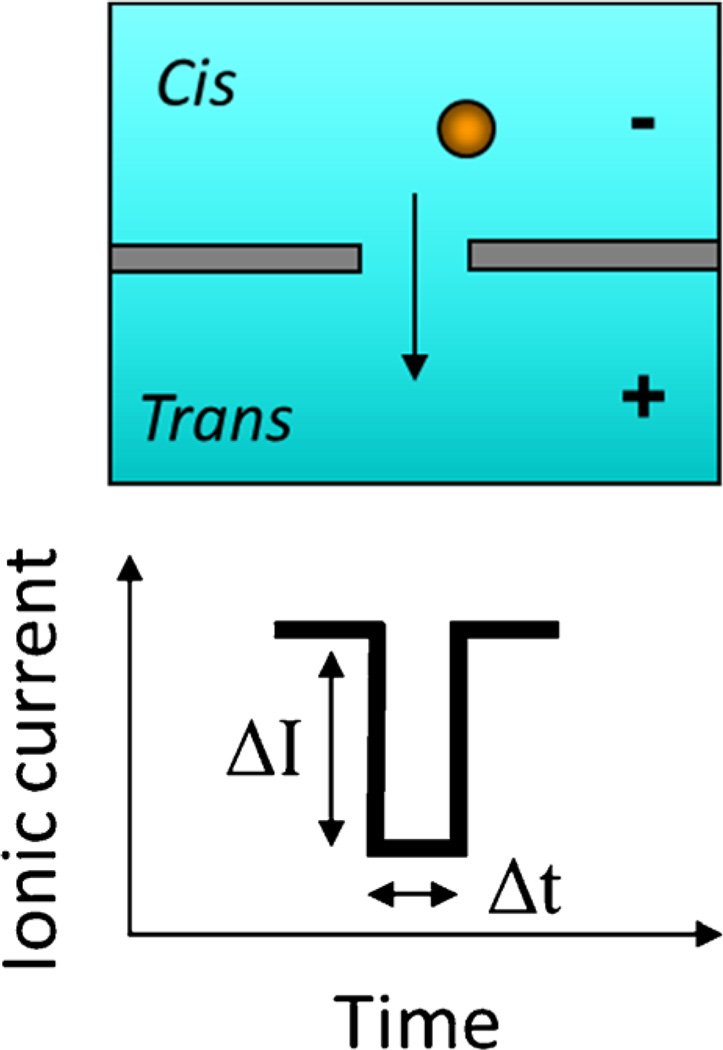
A solid-state nanopore, which utilizes a characteristic electrical signal for detection, is one of the powerful nanoscale structures which have promise for integration with optofluidics. A solid-state nanopore is a tiny opening in a thin inorganic membrane. Both sides of this membrane are filled with electrolyte solution. When a voltage is applied across this nanopore, an ionic current will be established through the nanopore. As a particle translocates from one side of the membrane to the other side through the nanopore, the nanopore will be blocked for a short period, during which a current drop can be recorded by a sensitive amplifier (Fig. 1). Theoretically, particles with different sizes, shapes, and electro chemical properties will generate current blockades with different amplitudes and durations (dwell times). Therefore, by analyzing the blockades, we can identify and distinguish different nanoparticles. Because of its extremely high sensitivity, a solid-state nanopore is a perfect tool for nanoparticle detection and a potential tool for nucleic acid sequencing. [23–31].
Fig. 1.
Schematic showing the detection principle for particles passing through a solid-state nanopore
Combination of solid-state nanopore and optofluidics
The combination of solid-state nanopore and optical detection is to some extent inevitable. Though substantial efforts and progress have been made to improve the performance, low spatial resolution due to fast particle translocation remains as one of the most important unresolved issues for solid-state nanopore technology. Limited detection bandwidth, flicker noise, and fast translocation speeds hinder the measurements of fine molecule structures. [32–35] Though modifying the properties of nanopores or the experimental parameters can somewhat address these issues, [36–38] researchers have also sought easier solutions from additional technologies. Fortunately, some developed optical detection methods, like fluorescence spectroscopy, have been demonstrated to be an effective approach to deal with the challenges that solid-state nanopores are facing.
Low noise from background signal is critical for optical detection of a single particle, so suppressing the background light in any optical detection scheme is hugely important. Great efforts have been made to address this issue for single molecule and bioparticle detection around nanopores, and several effective solutions have been developed. Total internal reflection fluorescence excitation can be used to collect optical signals from molecules near a liquid-membrane interface. [39] But the refractive index of the buffer in the trans chamber (Fig. 1) needs to be selected carefully to ensure the total internal reflection excitation. Confocal microscopy, which eliminates out of focus light, is another frequently used method for optical noise suppression. [40, 41] With the help of plasmonic effects, excitation light can be confined even more tightly. With light focused in a zeptoliter volume, the combination of zero-mode waveguide and solid-state nanopore has been applied for the detection of DNA translocation. [42, 43] Ca2+ sensitive dyes are also used for optical detection of DNA. [44, 45] The Ca2+ ion gradient across the nanopore ensures a strong fluorescence signal only within the region close to the nanopore. These successful demonstrations of electro-optical detection have proven the power of combining nanopores and optical detection. Furthermore, electro-optical detection of color-coded expanded-DNA has even demonstrated the ability to provide sufficient spatial resolution to distinguish four different nucleotides, suggesting a way towards potential electro-optical nucleotide sequencing with a solid-state nanopore. [46, 47].
Though careful alignment is needed, currently the top-down microscope (Fig. 2a) is still the most common way to couple light into and gather optical signals from the nanopore area. Depending on the application, different nanopore geometries may be required. A vertically oriented solid-state nanopore is good enough for single particle detection, but is not convenient for particle trajectory tracing. Recently horizontally oriented solid-state nanopores have been embedded into microfluidic devices (Fig. 2b), and these nanopore-optofluidic devices make the optical observation of particle movement easier. [48, 49] However, it is currently still difficult to control the shape of the horizontally oriented nanopore and to make its diameter as small as the vertical nanopore. Also the advantages of fluidics control have not been fully utilized. We can predict that after noting the benefits of the combination of nanopore and optical detection, the trend of making nanopore-optofluidic devices has only just begun.
Fig. 2.
Commonly used experimental setups for electro-optical detection with solid-state nanopore. a Vertical nanopore with top-down microscope. b Horizontal nanopore with top-down microscope
In the remainder of this article we will focus on the incorporation of solid-state nanopore into an ultrasensitive optofluidic platform, based on liquid-core anti-resonant reflecting optical waveguides (ARROWs), where the precise nanopore shape, flexible arrangement of waveguides and nanopore, and fluidics flow control are realized within a single device.
Highly sensitive optofluidic platform
Principle and fabrication methods
The beauty of optofluidics is the integration and interaction of light and fluid flow in the same device. Most of the optofluidic applications in bioanalysis rely on the interaction of light and fluids at an interface which is typically implemented with specific binding of a target to a solid surface and interaction with the evanescent tail of an optical mode that propagates through a solid waveguide. [10] This approach faces the challenges of efficiently transporting the target to the surface and utilizing only a small fraction of the input optical power for particle interaction.
An attractive alternative is, therefore, to implement sensing in flow which requires optical signals to reach and be transported in the fluidic channels themselves. Thus, the challenge is to develop a hollow optical waveguide which also allows liquid, whose refractive index is lower than the surrounding materials, to flow inside. Due to this index mismatch, conventional guiding by total internal reflection is not easily implemented for this purpose. Some approaches have been demonstrated, such as Teflon AF waveguides, liquid core liquid cladding waveguides, nanoporous cladding waveguides, slot waveguides, and photonic crystal fibers. [50–54] However, these solutions still have some limitations in terms of fabrication or integration.
One of the solutions to the above issue is to use anti-resonant reflecting optical waveguides (ARROWs). [55] When the thickness of each cladding layer is designed to meet the anti-resonant condition, low transmission through cladding layers can be achieved. As a result, the high reflectivity in the transverse direction will confine the light to the low-index liquid core. Optical connection between the liquid-core waveguide and the rest of the chip is accomplished using solid-core ARROW ridge waveguides that are parallel or vertically aligned with the liquid-core channel. [56] Fig. 3a(i) shows the original and typical layout that is used for single particle detection and particle manipulation. When applied to single particle fluorescence detection, excitation light is coupled into the solid-core waveguide from an optical fiber. When a target particle passes the excitation volume, the fluorescence from it will propagate along the liquid-core waveguide, be coupled into the connecting solid-core waveguide, and finally collected by a subsequent detection setup.
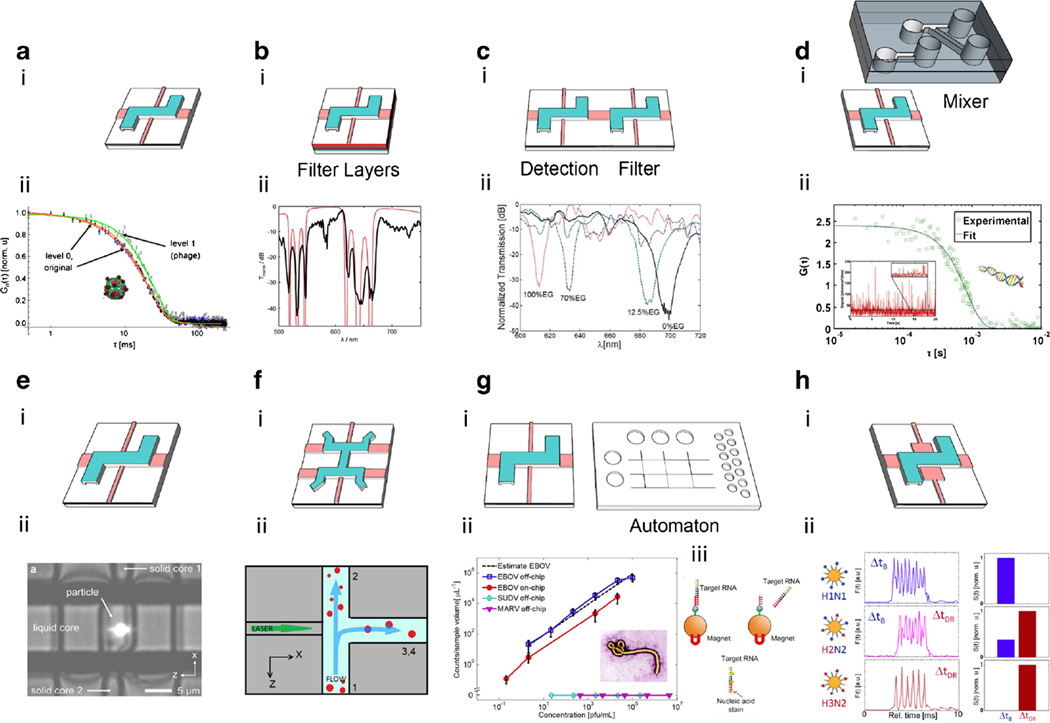
Fig. 3.
Variations of ARROWs and their applications. a i. Typical layout of ARROW-based optofluidic chip; ii. Fluorescence autocorrelation functions of Qβ bacteriophage. Original signal was separated into two levels. b i. ARROW with filter layers; ii. Optofluidic filter experimental (thick line) and calculated design (thin line) spectral response for a 4 mm long liquid-core ARROW waveguide. c i. Dual-channel chip with two different liquid-core sections for detection and tunable filtering; ii. Ethylene glycol (EG) spectral tuning. d i. PDMS mixer integration with an ARROW optofluidic chip; ii. Fluorescence autocorrelation function of detected λ-DNA molecules. e i. ARROW with offset solid-core waveguide; ii. Bright-field and fluorescence image of a trapped 1 µm bead. f i. ARROW with “H″ layout; ii. Principle of optofluidic particle sorting. g i. ARROW chip and PDMS based microfluidic automaton; ii. Concentration-dependent particle counts for off-chip (open squares) and using the automaton (solid circles) sample preparation; iii. The steps that were implemented on the automaton chip. h i. ARROW with MMI waveguide; ii. Two-color combinatorial detection of single viruses. Singly labeled viruses appear in only one Δt channel, whereas the double-labeled virus creates signal in both color channels
Liquid-core ARROWs have been being continuously optimized to diminish the waveguide loss, improve the sensitivity, and add new features. For example, single overcoat layers were introduced to increase the solid- to hollow-core waveguide coupling. [57] Self-aligned pedestals were used in the chip design to reduce the waveguide loss. [58] The background noise baseline above the detector dark counts of liquid-core waveguide was reduced by a factor of 10 by using sputtered Ta2O5 instead of vapor deposited SiN as a cladding layer. [59, 60] The optical properties of ARROWs can also be fine-tuned. Spectral filtering was implemented by tailoring three bottom layers of the liquid-core waveguide. [61] A dual-core configuration consisting of the standard analysis channel and a tunable filter section demonstrated its spectral tunability by tuning both core index and pH. [62] Excellent chip performance has led to numerous applications in high-sensitivity particle analysis.
Single particle detection
One of the most important applications of optofluidics is to detect single particles. Various particles have been detected and analyzed with ARROW based devices.
Single molecule detection with ARROW chips was first demonstrated by on-chip fluorescence detection and correlation spectroscopy (FCS) measurements of Alexa 647 dye molecules. [63, 64] Dye solutions with concentrations as low as 10pM were measured. Concentration values extracted from fits to the autocorrelation traces of the signal revealed that less than one (0.35) molecule on average was detected within the excitation volume. In the meantime, a modified autocorrelation function was proposed, which was later used for lots of ARROW-based on-chip single molecule detection. Shortly thereafter, ARROW devices showed their ability for high-performance bioanalysis. [65] FCS analysis down to 1.58 nM concentration of liposomes labeled with Alexa dye not only yielded a mean concentration of 0.64 liposomes in the excitation volume, but also demonstrated the ability to incorporate electrical bioparticle manipulation, here by electrokinetically driving the target particles through the fluidic channel. The velocity of liposomes was changed from around 100 µm/s to around 350 µm/s by adjusting the voltage across the liquid-core waveguide.
Later, Qβ bacteriophages were detected on the single virus level and analyzed using an ARROW chip. [66] The Qβ bacteriophages were labelled covalently with fluorescent dye molecules so that each of these 26 nm large spherical biomolecules was tagged with a couple hundred fluorophores. Diffusion coefficient, flow velocity and concentration of Qβ bacteriophage were extracted using FCS. A careful FCS analysis of the signals lead to the conclusion that a partial disassembly of the phage occurred. More complex methods, such as fluorescence cross-correlation spectroscopy (FCCS), were implemented on these chips as well. [67] Cy3 and Cy5 fluorescent dyes were placed at the same end of oligonucleotides after annealing to form a FRET (fluorescence resonance energy transfer) pair. As the temperature was increased, double-stranded DNA had dissociated into single stranded oligonucleotides and both FRET and FCCS signals decreased.
Particle manipulation
Besides single particle detection, the ARROW platform is also capable of manipulating and trapping particles. Different kinds of trapping methods were successfully realized on these devices. Particles can be trapped by a novel loss-based dual beam trap that is formed by the liquid-core waveguide loss resulting in decaying optical scattering force along the channel. Therefore two counter-propagating waves will create a potential well, where individual or groups of particles can be optically trapped, the latter enabling particle concentration in a small volume to enhance the signal for optical detection. As a demonstration, Escherichia coli attached to a latex particle was optically trapped and analyzed using this loss-based trap. [68, 69].
Yet another type of particle trap was enabled by using ARROWs with offset solid-core waveguides. [70] Two solid-core waveguides define two overlapping excitation volumes, the center of which is the equilibrium position. Deviation from this position results in modulations of the fluorescence signal that is used as feedback for electrokinetic forces that maintain the particle location at the equilibrium position. The use of electrokinetic forces allowed the trap to be operated at 105 times lower optical power. The bioanalysis application of this electro-optical trapping method was demonstrated by studying the DNA fluorescence dynamics in Escherichia coli bacteria. An E. coli bacterium was trapped, and the photobleaching process of stained DNA was observed simultaneously. In addition, in an “H″ shaped network of liquid-core waveguides, particles were sorted optically in a pressure-driven flow. [71] By tuning laser power and flow speed, all particles above a certain size can be sorted out from the stream optically. Most recently, multi-particle trapping was implemented on ARROW chips using the characteristic spot patterns produced by a multi-mode interferometer (MMI) waveguide section. [72].
Integration of functional add-ons
Vertical integration of other microfluidic devices brought the advantage of fluids control to the platform. The combination of a PDMS-based fluidic handling layer and silicon-based ARROW chips demonstrated the capabilities of mixing, distribution, and filtering with optical single particle detection. Combined labeling and detection of single λ–DNA molecules was accomplished in such a hybrid integrated device. [73] Beyond that, a specifically designed valve-controlled microfluidic layer (automaton) can carry out complex up-stream sample preparation steps. A hybrid automaton-ARROW device ingeniously combined sample preconcentration and direct detection of individual viral RNA molecules within a single device to quickly and accurately detect Ebola virus over more than seven orders of magnitude and down to 0.2 pfu/mL, covering the entire clinically relevant concentration range. [74] This achievement was also a first time demonstration of the possibility of replacing PCR techniques with a compact lab-on-a-chip system.
Very recently, one of the key concepts used in fiber optic communication, wavelength division multiplexing, was re-envisioned in the context of biomolecule analysis on an optofluidic ARROW chip. [75] A wide solid-core waveguide, acting as a single multimode interference (MMI) waveguide creates wavelength-dependent spot patterns along the intersecting liquid-core waveguide. When particles pass the spots, a series of fluorescence spikes can be detected. Since the spot width and spot spacing depend on the wavelength, this device enables the identification of differently labelled targets by deciphering the temporal fluorescence pattern. Identification of three kinds of single-color labelled and combinatorially labelled viruses was demonstrated, and more exciting applications with this technique can be expected.
Optofluidic chips containing integrated solid state nanopores
As stated in the introduction, integration of solid-state nanopore with optofluidics is still an ongoing area of development with many untapped and unoptimized opportunities. Many challenges can be resolved when the ARROW-based platform is chosen for the solid-state nanopore integration. First of all, the top of the liquid-core waveguide is made of SiN and SiO2, which are the materials usually chosen for solid-state nanopores. Since the solid-state nanopore will be vertically fabricated, all the techniques developed for solid-state nanopore fabrication can be directly applied. Optical alignment is also ensured easily and with high precision, since the light propagation is well defined by the waveguide network. Also, the planar configuration assures that there will be no false fluorescence signal, which means a particle that does not get into the liquid-core waveguide will not be excited accidently. Last but not least, the nanopore location along the liquid channel is flexible, allowing us to fulfill different detection purposes.
The combination of a solid-state nanopore with an ARROW chip merges two powerful single particle detection technologies, realizing a novel electro-optofluidic platform for bioanalysis. After elaborate efforts were made to optimize the fabrication methods of incorporating solid-state nanopores into ARROW devices, the opto-electrical single molecule analysis was demonstrated for the first time on an optically and fluidically integrated chip.
Methods for fabrication and their features
The ARROW chip poses unique nanopore fabrication challenges since the top of the liquid-core waveguide is several microns thick which results in unacceptably high aspect ratios for direct milling of a nanoscale opening. To address this problem, a two-step fabrication process was developed for defining nanopores in ARROW channels. [76] At any location along the liquid-core waveguide channel, a micropore of ~2–4 µm diameter is defined with a dry etch process. This etch is terminated by a metal etch stop layer placed on top of the first ~100 nm thick ARROW cladding layer. After removal of the etch stop layer, the nanopore is milled with a focused gallium ion beam.
In order to investigate how to optimize their dimensions, nanopores with different diameters and thicknesses have been tested and compared, with the aim of finding how the diameter and thickness influence the electrical signal. Many models have also been developed to simulate the behavior of solid state nanopores. [77–81] While the aperture size and thickness of the solid-state nanopore are probably the most important characteristics, the fabrication-dependent shape has a noticeable influence on its behavior as an electrical sensor. However, in most of the experiments or simulations, the solid-state nanopore was approximated simply as a regular cylindrical opening.
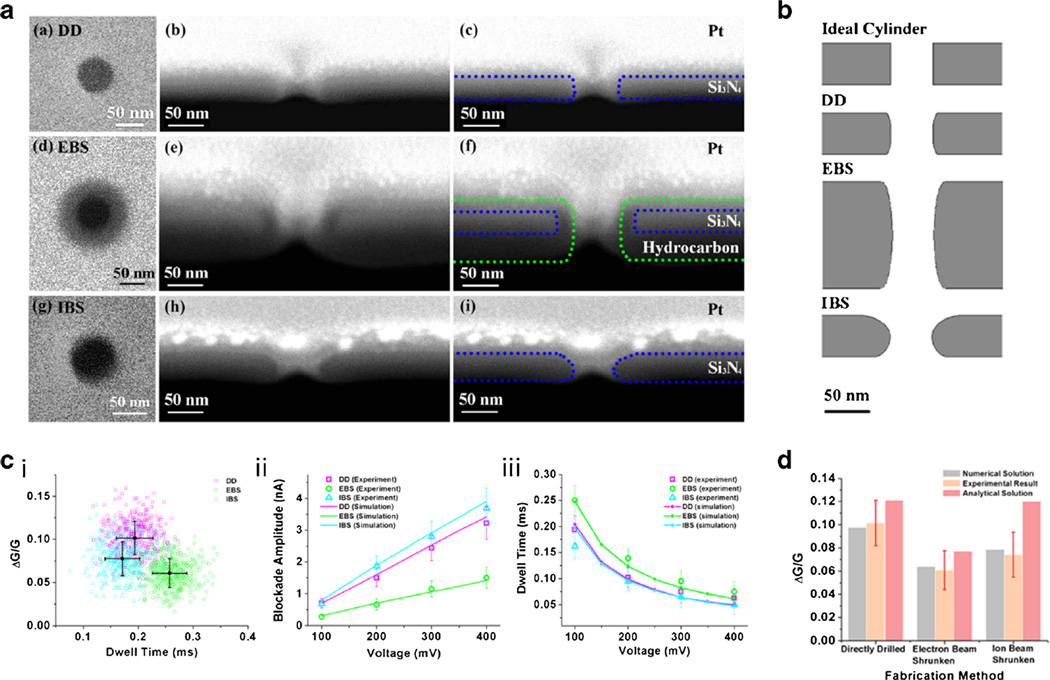
This correlation was investigated experimentally by defining nanopores of 50 nm diameter in three different ways: direct milling (DM), ion-beam (IB) and electron-beam (EB) assisted shrinking. [82] After single particle current blockade measurements were done with these nanopores, they were cross sectioned using a focused ion beam for shape characterization. At last, the simulated electrical behaviors based on these shapes were compared with the experimental results. The cross-sections of three types of nanopores were imaged and are shown in Fig. 4a. The shape of the DM nanopore is close to a cylinder, save for some rounding of the edges on the top and the bottom of the cylinder. The size of DM nanopore is also as expected, 50 nm wide and 50 nm long. The EB-shrunk nanopore has a cylindrical shape, too. However, after the EB scanning, deposition of hydrocarbons in the vacuum chamber thickens the membrane from 50 nm to 130 nm, thus elongating the nanopore significantly. The deposition of hydrocarbons was further confirmed by element-specific EDS spectroscopy. IB-shrunk nanopores, finally, deviate strongly from a cylindrical shape. Being close to a cone, they have a wider opening on the top and a narrower opening on the bottom. The resulting current blockades can be modeled very well. Figure 4c (i) shows the distribution of blockade depth and duration for the three pore types. Clear differences are observed despite their nominally identical aperture size. These differences are fully reproduced by taking into account the full 3D shape of the pore. Changes in blockade amplitude and dwell time under different voltages are compared using experimental and simulated results, and from Fig. 4c (ii–iii) we again find good agreement.
Fig. 4.
a Scanning electron micrographs of 50 nm nanopores made on 50 nm membranes. b The shapes of nanopores built in the model. c i. Scatter plot of events detected by three kinds of nanopores when the voltage is 100 mV; ii. The change of blockade amplitude versus the voltage; iii. Normalized experimental and simulated dwell time versus voltage. d The comparison between relative conductance changes predicted by numerical and analytical solutions, and experimental results. [84]
The most straightforward way to model the current blockade is to calculate the resistance change by calculating the excluded volume when a particle is present in the nanopore, while assuming the nanopore has a regular shape. Here, we further compared experimental results and our simulation with a recently corrected analytical solution for the relative conduction change: ΔG/G = d3/[(L+πD/4)D2], where G, d, L and D are nanopore conductance, particle diameter, nanopore length and nanopore diameter, respectively. [83].
In the analytical model, DM and IB shrunk nanopores with 50 nm diameter and 50 nm length were used, and EB shrunk nanopore with 50 nm diameter and 130 nm length was used. Compared to the numerical simulation, the analytical solution deviates more from the experimental data (Fig. 4d). The DM nanopore is the one that is closest to the ideal shape of a 50 nm wide and 50 nm long cylinder, thus the analytical solution can approximately predict the relative conductance change. When the shape of the nanopore deviates a lot from an ideal cylinder, the analytical solution gives a large difference between calculation and experimental data, which is the case of IB shrunk nanopores. Therefore, the numerical method is more powerful when an accurate estimation is needed for a nanopore with an irregular shape.
The nanopore as a smart gate
The first demonstration of incorporating electrical sensing of single biomolecules into optofluidic devices was given by detecting single ribosomes. [85] A nanopore with a 40 nm aperture was defined in an ARROW chip using ion-beam assisted shrinking as described above. The arrangement on the chip is schematically shown in Fig. 5a(i) along with a photograph of a nanopore-ARROW device (inset). A solution containing 50S ribosomal subunits was introduced into the fluid reservoir placed on top of the nanopore, and potentials on the order of 1 V were applied to move the ribosomes into the fluidic channel in single file. The observed electrical signal for different applied voltages is shown in Fig. 5a(ii); and the inset shows a characteristic blockade signal caused by a single ribosome. Blockades of ~4 % of the total ionic current were well resolved. Notably, the rate of translocations depends strongly on the applied potential as is shown in Fig. 5a(iii). The detection rate can be varied from thousands per seconds to zero over the span of ~250 mV. This strong dependence can be used in future devices to rapidly turn the nanopore gate on and off between experiments.
Fig. 5.
a i. Schematic view of intersecting solid-core (orange) and liquid-core (blue) optical waveguides on a silicon chip with particles and electrodes in reservoirs (inset: photograph of chip); ii. The observed electrical signal for different applied voltages; iii. Translocation rate versus applied voltage. b i. Fluorescent 100 nm and 200 nm nanoparticles are translocated through a 250 nm nanopore; ii. Electrical blockade (top) and fluorescence (bottom) signals showing correlated single-particle detection events; iii. Cross-correlation of electrical and optical signals; iv. Histogram of the optical brightness distribution for each particle type; v. Scatter plot of electrical blockades; vi. Multi-parameter analysis enabling assignment of optical properties to particle subpopulations. Dashed lines: optical signal range with ambiguous particle size assignment. c i. Schematic view of the virus/nanoparticle mixture and nanopore; ii. Electrical blockade (black) and spectrally resolved optical fluorescence signals from viruses (red) and nanoparticles (blue); iii. Scatter plot of electrical signals; iv. Cross-correlation of optical and electrical virus detection signals for various combinations of duration/spectral subpopulations. d i. Schematic view of the λ-DNA molecule and nanopore; ii. Electrical blockades (black) and optical signals (red); iii. The cross correlation function between electrical and optical signals; iv. Blockade amplitude versus voltage. Duration versus voltage. Capture rate versus voltage
Correlated electro-optical detection and discrimination of nanoparticles
As a proof of principle demonstration, correlated electro-optical detection of single 500 nm, 200 nm, and 100 nm nanoparticles were achieved, respectively. A nanopore with the size of 520 nm was used for measuring 500 nm nanoparticles. When 4 V voltage was applied between reservoirs 1 and 3, 500 nm nanoparticles were driven through the nanopore. When the nanoparticles got into the channel, they were immediately excited by the laser, resulting in clear, correlated fluorescence signals. Similarly, 200 nm and 100 nm nanoparticles were measured with 260 nm and 160 nm nanopores as well.
Next, a mixture of fluorescent nanoparticles of different diameter (100 and 200 nm) was introduced to a 250 nm wide nanopore. [86] Once translocated through the pore by a 3 V applied voltage, the particles were moved along the liquid-core ARROW channel with negative pressure. Figure 5b(ii) shows the resulting electrical and optical signals. In both cases, clean signatures from individual nanoparticles are observed with high signal-to-noise ratio. The strong correlation peak at 5.8 s confirms the correlated electro-optical detection of single nanoparticles, and suggests the flow velocity inside the liquid-core channel is 270 µm/s given a nanopore-excitation spacing of about 1.5 mm. Thus, the correlated single-particle measurements allow for immediate extraction of the flow speed in the ARROW channel.
It is expected that two kinds of nanoparticles can be distinguished using either electrical or optical signal. However, the bead subpopulations did not get resolved in the histogram of optical signal intensity, due to the variation of the exact locations of particles within the optical excitation volume. With the help of the solid-state nanopore, electrical blockades of two kinds of nanoparticles could be clearly identified due to their different amplitudes (Fig. 5b(v)). Because of the one-to-one correlation relationship, now the optical signal of each nanoparticle can be recognized with the help of the electrical signal. With this information in hand, each optical peak can be assigned to a particle type and a histogram of the optical brightness distribution for each particle type can be created (Fig. 5b(iv)). Both Fig. 5b(iv) and Fig. 5b(vi) suggest that within a certain intensity region the nanoparticles cannot be discriminated using the optical signal alone, verifying the advantage of the correlated electro-optical detection.
Detection and identification of viruses and λ-DNA molecules
A mixture of 100 nm fluorescent beads and ~100 nm large influenza (H1N1) viruses was investigated using a ~ 150 nm wide integrated nanopore. [86] The viruses were labeled to fluoresce in the red (~670 nm) while the bead fluorescence is in the blue, so the particles could be identified directly using the optical signals. The resulting electrical and optical signatures for this mixture are shown in Fig. 6c(ii). Again, clear blockades and optical spikes are resolved, representing the first demonstration of detection of single virus particles using two modalities on a single chip. Moreover, the single particle nature of the optical detection can be unambiguously proved as each particle has to transverse the nanopore in single file. Because of the similar sizes of two kinds of particles, analyzing the blockade amplitude alone will not help distinguish different particles. However, the data points naturally form two subpopulations with different durations, which are likely caused by different electrical properties of different particles. Now the clearly discriminated optical signals enable the distinct identification of the electrical blockades. The blockades were then separated into two subpopulations (blockades with short and long durations) with the dashed line in Fig. 6c(iii). The cross correlation functions between electrical blockades in two subpopulations (short and long) and optical signals in two channels (red and blue) immediately show that viruses (red) have long durations (>4 ms) while the nanoparticles (blue) have shorter (<4 ms) durations. These results prove again that individual labeled viruses can be counted and identified unambiguously from a particle mixture using the combination of electrical and optical signal channels.
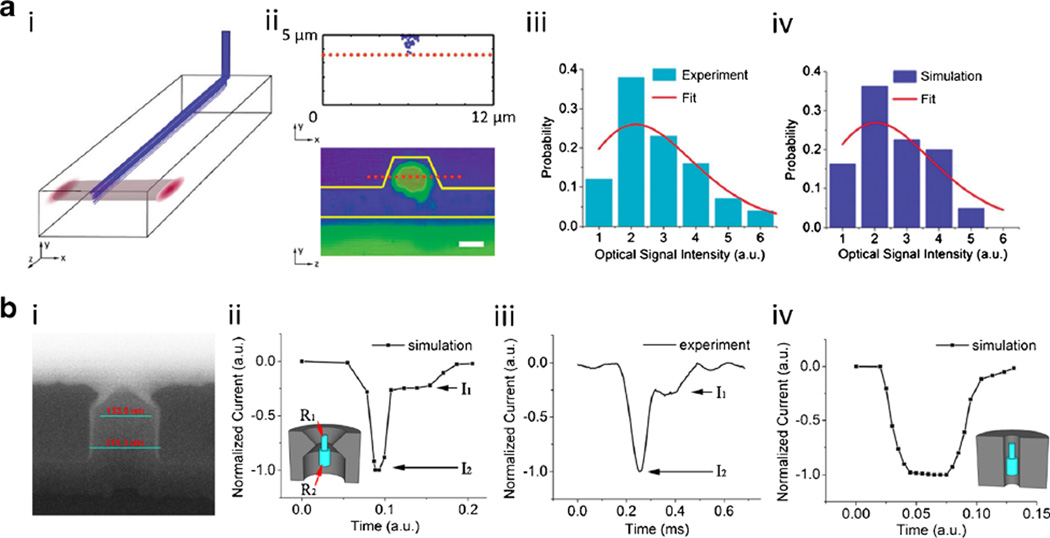
Fig. 6.
a i. The calculated trajectories of 100 particles in the liquid-core channel; ii. Top: a cross section view of the particle distribution. Bottom: the optical mode in the solid-core waveguide; iii. Experimental intensity distribution (line: Poisson fit); iv. Simulated intensity distribution when center of the optical mode is located 3.6 µm above the bottom of the channel (line: Poisson fit). b i. The cross section image of a nanopore shrunk by SiO2 deposition; ii. A simulated blockade caused by a particle going through a nanopore with pointy edge. Inset: the structures of nanopore and particle; iii. An experimentally observed blockade; iv. A simulated blockade caused by a particle going through a cylindrical nanopore. Inset: the structures of nanopore and particle
The solid-state nanopore has been considered as a potential tool for next generation nucleic acid sequencing, and ARROW chips have been demonstrated to be sensitive enough to detect single DNA molecules. [74] Therefore, the next step was to prove the feasibility and the advantages of the integrated nanopore-ARROW device for single DNA molecule detection. λ-DNA molecules were introduced into the liquid-core channel through a 20 nm wide nanopore. [87] Under voltages of 4.5 V, 5.4 V and 6.3 V, clear electrical blockades were observed. By analyzing the trends of blockade amplitude and duration changes, information about the trans-location dynamics can be revealed (Fig. 6d(iv)). The average blockade amplitude increases linearly with incremental voltage, following the linear fitting function at a rate of 540 pA/V. The x-intercept of 2.8 V suggests there is a diffusion limited capture region above a threshold voltage of 2.8 V. [88] The duration decreases exponentially with the voltage, pointing towards interactions between the molecules and the nanopore. [89] A linear dependence of the capture rate on rising voltage across the nanopore with a rate of 0.1 s−1 V−1 indicates the capture process is governed by thermal diffusion. [90] Again, the cross correlation function between electrical and optical signals shows a strong cross correlation peak at 0.18 s (Fig. 6d(iii)), indicating λ-DNA travels with a rather uniform velocity of 8.3 mm/s between the nanopore and the waveguide intersection.
Particle trajectory simulation and particle location determination
The optical signal is not just correlated with the corresponding electrical signal and flow velocity, but also the location of the particles in the optical excitation area. After particles are loaded into the channel, where they are interacting with the exciting light is of great interest and importance, since it is potentially helpful for more efficient optical detection in the future. The mode location of the excitation light can always be acquired by taking a mode image at the end of output solid-core waveguide; however, where the optical mode is located inside the liquid-core channel cannot be acquired directly without destroying the chip.
Figure 6a(iii) shows the intensity distribution of optical signals obtained from the experiment. A Poisson distribution fitting reveals the experimental expected intensity is 2.7. In order to simulate the particle distribution in the ARROW device, a 3D model consisting of a liquid core channel and a nanopore inlet is built with COMSOL. Laminar flow in the channel is simulated based on the Navier-Stokes flow. 100 Newtonian particles are introduced through the nanopore inlet. The positions of the particles at the excitation area are obtained from the simulations and imported in Matlab. Optical excitation and collection mode profiles are combined to obtain the signal intensity of each particle, depending on its position. At last, a histogram plot is created based on the intensity statistics. A good match between experimental and simulated intensity distributions will not be found until the optical mode is 3.6 µm high (Fig. 6a(iv)). Most importantly, this analysis serves as proof that the intensity variation in the optical detection of single particles in an ARROW device is due to fluctuations in particle locations and not other effects such as non-uniform labeling or particle aggregation.
Sensitivity enhancement by tuning nanopore shape
The shape of a nanopore shrunk by FIB-assisted SiO2 deposition was characterized. It can be seen from Fig. 6b(i) that a thin layer of deposited SiO2 protrudes from the top of the initial nanopore and forms a pointed end. In order to simulate what kind of electrical blockade we can get from a nanopore with such a cross section shape, a structure with a similar shape was modelled in COMSOL (inset in Fig. 6b(ii)). The minimum diameter of the structure was 20 nm. The blue particle formed by two connected cylinders is meant to imitate a folded DNA molecule. The radius of the thicker cylinder is twice as large as that of the thinner cylinder (R2 = 2R1). After moving the particle through the nanopore and calculating the ionic current at each point, we got a step-like blockade (Fig. 6b(ii)). We also found that the ratio between the amplitudes of two steps equals the ratio of the cross section areas of two cylinders (I2/I1 = πR22/πR12 = 4). However, when the same particle was moved through a regular cylinder, step-like blockades are not observed, indicating that the fine structure was not detected. The cross section analysis and the simulation suggest that the nanopore sensitivity can be enhanced by using a nanopore shrunk by SiO2 deposition due to the introduction of a limiting aperture that is very sharply defined in the vertical direction. The simulation matches well with some experimentally observed blockades, as seen in Fig. 6b(iii). Because I2/I1 ≈ 4, based on the above discussion, we can deduce it could be caused by a folded λ-DNA molecule of which the thicker part is twice as thick as the thinner part.
Summary and outlook
In this paper, we reviewed the current progress of the combination of solid-state nanopore and optofluidics, especially the integration of solid-state nanopores into ARROW-based optofluidic devices. The capabilities of this liquid-core waveguide platform were illustrated in detail, and various applications in single particle detection and particle manipulation were discussed. After the fabrication methods of forming solid-state nanopores into ARROW chips were investigated and compared, we presented a series of electro-optical detection examples on nanopore-ARROW devices. Single polystyrene nanoparticles, H1N1 viruses, and λ-DNA were successfully detected simultaneously with ionic current blockades and fluorescence signals. Moreover, information about the fluid flow and molecular configurations were extracted by using both electrical and optical signals. We also proposed and demonstrated that the sensitivity of the device can be further improved by using a SiO2 deposition-shrunk nanopore. Nanopores in optofluidic chips face the same challenges as solid-state nanopores used purely for electrical sensing. Care has to be taken to minimize electrical noise. The fabrication of extremely small apertures on the order of ~2 nm for advanced single nucleic acid analysis is challenging, requiring thin membranes and exquisite control over the milling and reshaping process. Moreover, the incorporation of nanopores with microfluidic channels requires additional considerations, such as balancing the impedance of the narrow, but short nanopore with that of the wider, but much longer fluidic channel. Finally, care has to be taken to design the nanopore placement and interface in such a way that the desired optical properties of the device are not compromised.
Although the sensitive electro-optical detection of single nanoparticles have been demonstrated, the application of nanopore-optofluidics in bioanalysis has just begun. In the near future, the expansion of this approach to incorporate feedback control is compelling. Deterministic introduction of one, and only one nanoparticle into the fluidic channel at a time can be accomplished by adjusting the potential across the pore upon translocation. When combined with advanced particle trapping capabilities, [69, 70] such a gated nanopore device can form the basis of a simple to use, high throughput instrument for single molecule studies. Moreover, the electrical signal provided by the nanopore can be used as a decision tool for subsequent routing and processing of the translocated particles in the optofluidic channel network. As both nanopore and optofluidic technology continue to mature, more novel and exciting applications will emerge.
Acknowledgments
We thank D.W. Deamer and H.F. Noller for fruitful discussions, T.D. Yuzvinsky for EDS spectroscopy analysis, Y. Zhao and T.A. Wall for ARROW chips fabrication, D. Ozcelik for particle trajectory simulations, and J.W. Parks for virus sample preparation. We acknowledge support by the W.M. Keck Center for Nanoscale Optofluidics at University of California, Santa Cruz, the NSF under grants CBET-1402848 and CBET-1159423, and the NIH under grants R01EB006097 and R21EB008802.
Contributor Information
Shuo Liu, Email: sliu@soe.ucsc.edu.
Holger Schmidt, Email: hschmidt@soe.ucsc.edu.
References
- 1.Stone HA, Stroock AD, Ajdari A. Engineering flows in small devices. Annu Rev Fluid Mech. 2004;36:381–411. [Google Scholar]
- 2.Squires TM, Quake SR. Microfluidics: fluid physics at the nanoliter scale. Rev Mod Phys. 2005;77:977–1026. [Google Scholar]
- 3.Beebe DJ, Mensing GA, Walker GM. Physics and applications of microfluidics in biology. Annu Rev Biomed Eng. 2002;4:261–286. doi: 10.1146/annurev.bioeng.4.112601.125916. [DOI] [PubMed] [Google Scholar]
- 4.Whitesides GM. The origins and the future of microfluidics. Nature. 2006;442:368–373. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 5.Dittrich PS, Manz A. Lab-on-a-chip: microfluidics in drug discovery. Nat Rev Drug Discov. 2006;5:210–218. doi: 10.1038/nrd1985. [DOI] [PubMed] [Google Scholar]
- 6.Psaltis D, Quake SR, Yang C. Developing optofluidic technology through the fusion of microfluidics and optics. Nature. 2006;442:381–386. doi: 10.1038/nature05060. [DOI] [PubMed] [Google Scholar]
- 7.Monat C, Domachuk P, Eggleton BJ. Integrated optofluidics: a new river of light. Nat Photonics. 2007;1:106–114. [Google Scholar]
- 8.Schmidt H, Hawkins AR. The photonic integration of non-solid media using optofluidics. Nat Photonics. 2011;5:598–604. [Google Scholar]
- 9.Erickson D, Sinton D, Psaltis D. Optofluidics for energy applications. Nat Photonics. 2011;5:583–590. [Google Scholar]
- 10.Fan X, White IM. Optofluidic microsystems for chemical and biological analysis. Nat Photonics. 2011;5:591–597. doi: 10.1038/nphoton.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt H, Hawkins AR. Optofluidic waveguides: I. Concepts and implementations. Microfluid Nanofluid. 2008;4:3–16. doi: 10.1007/s10404-007-0199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hawkins AR, Schmidt H. Optofluidic waveguides: II. Fabrication and structures. Microfluid Nanofluid. 2007;4:17–32. doi: 10.1007/s10404-007-0194-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim S, Streets AM, Lin RR, Quake SR, Weiss S, Majumdar DS. High-throughput single-molecule optofluidic analysis. Nat Methods. 2011;8:242–245. doi: 10.1038/nmeth.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coskun AF, Cetin AE, Galarreta BC, Alvarez DA, Altug H, Ozcan A. Lensfree optofluidic plasmonic sensor for real-time and label-free monitoring of molecular binding events over a wide field-of-view. Sci Rep. 2014;4:6789. doi: 10.1038/srep06789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bertucci A, Manicardi A, Candiani A, Giannetti S, Cucinotta A, Spoto G, Konstantaki M, Pissadakis S, Selleri S, Corradini R. Detection of unamplified genomic DNA by a PNA-based microstructured optical fiber (MOF) Bragg-grating optofluidic system. Biosens Bioelectron. 2015;63:248–254. doi: 10.1016/j.bios.2014.07.047. [DOI] [PubMed] [Google Scholar]
- 16.Chow E, Grot A, Mirkarimi LW, Sigalas M, Girolami G. Ultracompact biochemical sensor built with two-dimensional photonic crystal microcavity. Opt Lett. 2004;29:1093–1095. doi: 10.1364/ol.29.001093. [DOI] [PubMed] [Google Scholar]
- 17.Liu P, Huang H, Cao T, Tang Z, Liu X, Qi Z, Ren M, Wu H. An optofluidics biosensor consisted of high-finesse fabry-pérot resonator and micro-fluidic channel. Appl Phys Lett. 2012;100:233705. [Google Scholar]
- 18.Sun Y, Fan X. Optical ring resonators for biochemical and chemical sensing. Anal Bioanal Chem. 2011;399:205–211. doi: 10.1007/s00216-010-4237-z. [DOI] [PubMed] [Google Scholar]
- 19.Fenzl C, Hirsch T, Wolfbeis OS. Photonic crystals for chemical sensing and biosensing. Angew Chem Int Ed. 2014;53:3318–3335. doi: 10.1002/anie.201307828. [DOI] [PubMed] [Google Scholar]
- 20.Deng Y-L, Juang Y-J. Electrokinetic trapping and surface enhanced Raman scattering detection of biomolecules using optofluidic device integrated with a microneedles array. Biomicrofluidics. 2013;7:014111. doi: 10.1063/1.4793224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song W, Psaltis D. Electrically tunable optofluidic light switch for reconfigurable solar lighting. Lab Chip. 2013;13:2708–2713. doi: 10.1039/c3lc50204j. [DOI] [PubMed] [Google Scholar]
- 22.Soltani M, Lin J, Forties RA, Inman JT, Saraf SN, Fulbright RM, Lipson M, Wang MD. Nanophotonic trapping for precise manipulation of biomolecular arrays. Nat Nanotechnol. 2014;9:448–452. doi: 10.1038/nnano.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dekker C. Solid-state nanopores. Nat Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 24.Stoloff DH, Wanunu M. Recent trends in nanopores for biotechnology. Curr Opin Biotechnol. 2013;24:699–704. doi: 10.1016/j.copbio.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahrami A, Doğan F, Japrung D, Albrecht T. Solid-state nanopores for biosensing with submolecular resolution. Biochem Soc Trans. 2012;40:624–628. doi: 10.1042/BST20120121. [DOI] [PubMed] [Google Scholar]
- 26.Feng Y, Zhang Y, Ying C, Wang D, Du C. Nanopore-based fourth-generation DNA sequencing technology. Genomics Proteomics Bioinformatics. 2015;13:4–16. doi: 10.1016/j.gpb.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miles BN, Ivanov AP, Wilson KA, Doğan F, Japrung D, Edel JB. Single molecule sensing with solid-state nanopores: novel materials, methods, and applications. Chem Soc Rev. 2012;42:15–28. doi: 10.1039/c2cs35286a. [DOI] [PubMed] [Google Scholar]
- 28.Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nat Nanotechnol. 2011;6:615–624. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 29.Zaino LP, Ma C, Bohn PW. Nanopore-enabled electrode arrays and ensembles. Microchim Acta. 2015 [Google Scholar]
- 30.Shi J, Hou J, Fang Y. Recent advances in nanopore-based nucleic acid analysis and sequencing. Microchim Acta. 2015:1–15. [Google Scholar]
- 31.Li J, Yu D, Zhao Q. Solid-state nanopore-based DNA single molecule detection and sequencing. Microchim Acta. 2015:1–13. [Google Scholar]
- 32.Uram JD, Ke K, Mayer M. Noise and bandwidth of current recordings from submicrometer pores and nanopores. ACS Nano. 2008;2:857–872. doi: 10.1021/nn700322m. [DOI] [PubMed] [Google Scholar]
- 33.Smeets RMM, Keyser UF, Dekker NH, Dekker C. Noise in solid-state nanopores. Proc Natl Acad Sci. 2008;105:417–421. doi: 10.1073/pnas.0705349105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smeets RMM, Dekker NH, Dekker C. Low-frequency noise in solid-state nanopores. Nanotechnology. 2009;20:095501. doi: 10.1088/0957-4484/20/9/095501. [DOI] [PubMed] [Google Scholar]
- 35.Tabard-Cossa V, Trivedi D, Wiggin M, Jetha NN, Marziali A. Noise analysis and reduction in solid-state nanopores. Nanotechnology. 2007;18:305505. [Google Scholar]
- 36.Yusko EC, Johnson JM, Majd S, Prangkio P, Rollings RC, Li J, Yang J, Mayer M. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat Nanotechnol. 2011;6:253–260. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fologea D, Uplinger J, Thomas B, McNabb DS, Li J. Slowing DNA translocation in a solid-state nanopore. Nano Lett. 2005;5:1734–1737. doi: 10.1021/nl051063o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mirsaidov U, Comer J, Dimitrov V, Aksimentiev A, Timp G. Slowing the translocation of double-stranded DNA using a nanopore smaller than the double helix. Nanotechnology. 2010;21:395501. doi: 10.1088/0957-4484/21/39/395501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soni GV, Singer A, Yu Z, Sun Y, McNally B, Meller A. Synchronous optical and electrical detection of biomolecules traversing through solid-state nanopores. Rev Sci Instrum. 2010;81:014301. doi: 10.1063/1.3277116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chansin GAT, Mulero R, Hong J, Kim MJ, deMello AJ, Edel JB. Single-molecule spectroscopy using nanoporous membranes. Nano Lett. 2007;7:2901–2906. doi: 10.1021/nl071855d. [DOI] [PubMed] [Google Scholar]
- 41.Kurz V, Nelson EM, Shim J, Timp G. Direct visualization of single-molecule translocations through synthetic nanopores comparable in size to a molecule. ACS Nano. 2013;7:4057–4069. doi: 10.1021/nn400182s. [DOI] [PubMed] [Google Scholar]
- 42.Auger T, Mathé J, Viasnoff V, Charron G, Di Meglio JM, Auvray L, Montel F. Zero-mode waveguide detection of flow-driven DNA translocation through nanopores. Phys Rev Lett. 2014;113:028302. doi: 10.1103/PhysRevLett.113.028302. [DOI] [PubMed] [Google Scholar]
- 43.Larkin J, Foquet M, Turner SW, Korlach J, Wanunu M. Reversible positioning of single molecules inside zero-mode waveguides. Nano Lett. 2014;14:6023–6029. doi: 10.1021/nl503134x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson BN, Assad ON, Gilboa T, Squires AH, Bar D, Meller A. Probing solid-state nanopores with light for the detection of unlabeled analytes. ACS Nano. 2014;8:11836–11845. doi: 10.1021/nn505545h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivankin A, Henley RY, Larkin J, Carson S, Toscano ML, Wanunu M. Label-free optical detection of biomolecular translocation through nanopore arrays. ACS Nano. 2014;8:10774–10781. doi: 10.1021/nn504551d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McNally B, Singer A, Yu Z, Sun Y, Weng Z, Meller A. Optical recognition of converted DNA nucleotides for single-molecule DNA sequencing using nanopore arrays. Nano Lett. 2010;10:2237–2244. doi: 10.1021/nl1012147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Assad ON, Di Fiori N, Squires AH, Meller A. Two color DNA barcode detection in photoluminescence suppressed silicon nitride nanopores. Nano Lett. 2015;15:745–752. doi: 10.1021/nl504459c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yukimoto N, Tsutsui M, He Y, Shintaku H, Tanaka S, Kawano S, Kawai T, Taniguchi M. Tracking single-particle dynamics via combined optical and electrical sensing. Sci Rep. 2013;3:1855. doi: 10.1038/srep01855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Angeli E, Volpe A, Fanzio P, Repetto L, Firpo G, Guida P, Lo Savio R, Wanunu M, Valbusa U. Simultaneous electro-optical tracking for nanoparticle recognition and counting. Nano Lett. 2015;15:5696–5701. doi: 10.1021/acs.nanolett.5b01243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Datta A, Eom I-Y, Dhar A, Kuban P, Manor R, Ahmad I, Gangopadhyay S, Dallas T, Holtz M, Temkin H, Dasgupta PK. Microfabrication and characterization of Teflon AF-coated liquid core waveguide channels in silicon. IEEE Sensors J. 2003;3:788–795. [Google Scholar]
- 51.Wolfe DB, Conroy RS, Garstecki P, Mayers BT, Fischbach MA, Paul KE, Prentiss M, Whitesides GM. Dynamic control of liquid-core/liquid-cladding optical waveguides. Proc Natl Acad Sci U S A. 2004;101:12434–12438. doi: 10.1073/pnas.0404423101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Risk W, Kim H, Miller R, Temkin H, Gangopadhyay S. Optical waveguides with an aqueous core and a low-index nanoporous cladding. Opt Express. 2004;12:6446–6455. doi: 10.1364/opex.12.006446. [DOI] [PubMed] [Google Scholar]
- 53.Almeida VR, Xu Q, Barrios CA, Lipson M. Guiding and confining light in void nanostructure. Opt Lett. 2004;29:1209–1211. doi: 10.1364/ol.29.001209. [DOI] [PubMed] [Google Scholar]
- 54.Mandal S, Erickson D. Optofluidic transport in liquid core waveguiding structures. Appl Phys Lett. 2007;90:184103. [Google Scholar]
- 55.Duguay MA, Kokubun Y, Koch TL, Pfeiffer L. Antiresonant reflecting optical waveguides in SiO2-Si multilayer structures. Appl Phys Lett. 1986;49:13–15. [Google Scholar]
- 56.Schmidt H, Yin D, Barber JP, Hawkins AR. Hollow-core waveguides and 2-D waveguide arrays for integrated optics of gases and liquids. IEEE J Sel Top Quantum Electron. 2005;11:519–527. [Google Scholar]
- 57.Phillips BS, Jenkins MH, Liu S, Schmidt H, Hawkins AR. Selective thin-film deposition for optofluidic platforms with optimized transmission. IEEE Photon Technol Lett. 2011;23:721–723. [Google Scholar]
- 58.Lunt EJ, Wu B, Keeley JM, Measor P, Schmidt H, Hawkins AR. Hollow ARROW waveguides on self-aligned pedestals for improved geometry and transmission. IEEE Photon Technol Lett. 2010;22:1147–1149. doi: 10.1109/LPT.2010.2051145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y, Jenkins M, Measor P, Leake K, Liu S, Schmidt H, Hawkins AR. Hollow waveguides with low intrinsic photoluminescence fabricated with Ta2O5 and SiO2 films. Appl Phys Lett. 2011;98:091104. doi: 10.1063/1.3561749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holmes MR, Liu S, Keeley J, Jenkins M, Leake K, Schmidt H, Hawkins AR. Hollow waveguides with low intrinsic photoluminescence fabricated with PECVD silicon nitride and silicon dioxide films. IEEE Photon Technol Lett. 2011;23:1466–1468. [Google Scholar]
- 61.Measor P, Phillips BS, Chen A, Hawkins AR, Schmidt H. Tailorable integrated optofluidic filters for biomolecular detection. Lab Chip. 2011;11:899–904. doi: 10.1039/c0lc00496k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ozcelik D, Phillips BS, Parks JW, Measor P, Gulbransen D, Hawkins AR, Schmidt H. Dual-core optofluidic chip for independent particle detection and tunable spectral filtering. Lab Chip. 2012;12:3728–3733. doi: 10.1039/c2lc40700k. [DOI] [PubMed] [Google Scholar]
- 63.Yin D, Deamer DW, Schmidt H, Barber JP, Hawkins AR. Single-molecule detection sensitivity using planar integrated optics on a chip. Opt Lett. 2006;31:2136–2138. doi: 10.1364/ol.31.002136. [DOI] [PubMed] [Google Scholar]
- 64.Yin D, Lunt EJ, Barman A, Hawkins AR, Schmidt H. Microphotonic control of single molecule fluorescence correlation spectroscopy using planar optofluidics. Opt Express. 2007;15:7290–7295. doi: 10.1364/oe.15.007290. [DOI] [PubMed] [Google Scholar]
- 65.Yin D, Lunt EJ, Rudenko MI, Deamer DW, Hawkins AR, Schmidt H. Planar optofluidic chip for single particle detection, manipulation, and analysis. Lab Chip. 2007;7:1171–1175. doi: 10.1039/b708861b. [DOI] [PubMed] [Google Scholar]
- 66.Rudenko MI, Kühn S, Lunt EJ, Deamer DW, Hawkins AR, Schmidt H. Ultrasensitive Qβ phage analysis using fluorescence correlation spectroscopy on an optofluidic chip. Biosens Bioelectron. 2009;24:3258–3263. doi: 10.1016/j.bios.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen A, Eberle MM, Lunt EJ, Liu S, Leake K, Rudenko MI, Hawkins AR, Schmidt H. Dual-color fluorescence cross-correlation spectroscopy on a planar optofluidic chip. Lab Chip. 2011;11:1502–1506. doi: 10.1039/c0lc00401d. [DOI] [PubMed] [Google Scholar]
- 68.Kühn S, Lunt EJ, Phillips BS, Hawkins AR, Schmidt H. Optofluidic particle concentration by a long-range dual-beam trap. Opt Lett. 2009;34:2306–2308. doi: 10.1364/ol.34.002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kühn S, Measor P, Lunt EJ, Phillips BS, Deamer DW, Hawkins AR, Schmidt H. Loss-based optical trap for on-chip particle analysis. Lab Chip. 2009;9:2212–2216. doi: 10.1039/b900555b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kühn S, Phillips BS, Lunt EJ, Hawkins AR, Schmidt H. Ultralow power trapping and fluorescence detection of single particles on an optofluidic chip. Lab Chip. 2010;10:189–194. doi: 10.1039/b915750f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leake KD, Phillips BS, Yuzvinsky TD, Hawkins AR, Schmidt H. Optical particle sorting on an optofluidic chip. Opt Express. 2013;21:32605–32610. doi: 10.1364/OE.21.032605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leake KD, Olson MAB, Ozcelik D, Hawkins AR, Schmidt H. Spectrally reconfigurable integrated multi-spot particle trap. Opt Lett. 2015;40:5435–5438. doi: 10.1364/OL.40.005435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Parks JW, Cai H, Zempoaltecatl L, Yuzvinsky TD, Leake K, Hawkins AR, Schmidt H. Hybrid optofluidic integration. Lab Chip. 2013;13:4118–4123. doi: 10.1039/c3lc50818h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cai H, Parks JW, Wall TA, Stott MA, Stambaugh A, Alfson K, Griffiths A, Mathies RA, Carrion R, Patterson JL, Hawkins AR, Schmidt H. Optofluidic analysis system for amplification-free, direct detection of ebola infection. Sci Rep. 2015;5:14494. doi: 10.1038/srep14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ozcelik D, Parks JW, Wall TA, Stott MA, Cai H, Parks JW, Hawkins AR, Schmidt H. Optofluidic wavelength division multiplexing for single-virus detection. Proc Natl Acad Sci. 2015;112:12933–12937. doi: 10.1073/pnas.1511921112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Holmes MR, Shang T, Hawkins AR, Rudenko M, Measor P, Schmidt H. Micropore and nanopore fabrication in hollow antiresonant reflecting optical waveguides. J Micronanolithography MEMS MOEMS JM3. 2010;9:023004. doi: 10.1117/1.3378152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao Q, Sigalov G, Dimitrov V, Dorvel B, Mirsaidov U, Sligar S, Aksimentiev A, Timp G. Detecting SNPs using a synthetic nanopore. Nano Lett. 2007;7:1680–1685. doi: 10.1021/nl070668c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matysiak S, Montesi A, Pasquali M, Kolomeisky AB, Clementi C. Dynamics of polymer translocation through nanopores: theory meets experiment. Phys Rev Lett. 2006;96:118103. doi: 10.1103/PhysRevLett.96.118103. [DOI] [PubMed] [Google Scholar]
- 79.Ghosal S. Electrokinetic-flow-induced viscous drag on a tethered DNA inside a nanopore. Phys Rev E. 2007;76:061916. doi: 10.1103/PhysRevE.76.061916. [DOI] [PubMed] [Google Scholar]
- 80.Tian P, Smith GD. Translocation of a polymer chain across a nanopore: a Brownian dynamics simulation study. J Chem Phys. 2003;119:11475–11483. [Google Scholar]
- 81.Meller A, Nivon L, Branton D. Voltage-driven DNA trans-locations through a nanopore. Phys Rev Lett. 2001;86:3435–3438. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 82.Liu S, Yuzvinsky TD, Schmidt H. Effect of fabrication-dependent shape and composition of solid-state nanopores on single nanoparticle detection. ACS Nano. 2013;7:5621–5627. doi: 10.1021/nn4020642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Davenport M, Healy K, Pevarnik M, Teslich N, Cabrini S, Morrison AP, Siwy ZS, Létant SE. The role of pore geometry in single nanoparticle detection. ACS Nano. 2012;6:8366–8380. doi: 10.1021/nn303126n. [DOI] [PubMed] [Google Scholar]
- 84.Liu S. Electro-opto-fluidics: nanopore-gated devices for multimodal analysis of single biomolecules. UNIVERSITY OF CALIFORNIA, SANTA CRUZ. 2014 [Google Scholar]
- 85.Rudenko MI, Holmes MR, Ermolenko DN, Lunt EJ, Gerhardt S, Noller HF, Deamer DW, Hawkins A, Schmidt H. Controlled gating and electrical detection of single 50S ribosomal subunits through a solid-state nanopore in a microfluidic chip. Biosens Bioelectron. 2011;29:34–39. doi: 10.1016/j.bios.2011.07.047. [DOI] [PubMed] [Google Scholar]
- 86.Liu S, Zhao Y, Parks JW, Deamer DW, Hawkins AR, Schmidt H. Correlated electrical and optical analysis of single nanoparticles and biomolecules on a nanopore-gated optofluidic chip. Nano Lett. 2014;14:4816–4820. doi: 10.1021/nl502400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu S, Wall TA, Ozcelik D, Parks JW, Hawkins AR, Schmidt H. Electro-optical detection of single λ-DNA. Chem Commun. 2015;51:2084–2087. doi: 10.1039/c4cc07591a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Heng JB, Aksimentiev A, Ho C, Marks P, Grinkova YV, Sligar S, Schulten K, Timp G. Stretching DNA using the electric field in a synthetic nanopore. Nano Lett. 2005;5:1883–1888. doi: 10.1021/nl0510816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu L, Liu H, Zhao W, Wang L, Hou C, Liu Q, Lu Z. Electrically facilitated translocation of protein through solid nanopore. Nanoscale Res Lett. 2014;9:140. doi: 10.1186/1556-276X-9-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wanunu M, Morrison W, Rabin Y, Grosberg AY, Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat Nanotechnol. 2010;5:160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]