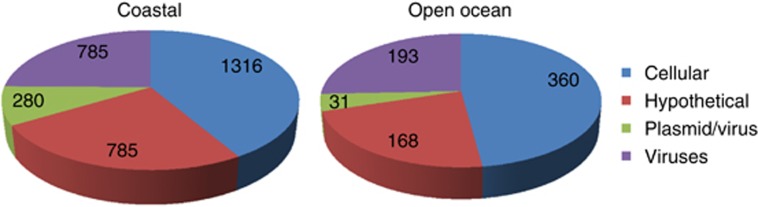
Figure 2.
Evaluation of the origin of MV-associated DNA sequences from coastal (left) and open ocean (right) samples. The original data on the best-blast hits and functional annotation of the DNA sequences recovered from marine MVs were obtained from the publication of Biller et al (2014). Sequences were categorized into four different groups according to the proteins they encode: (i) cellular proteins (bacterial, archaea and eukaryotic proteins that are rarely found in viruses and plasmids); (ii) hypothetical proteins (proteins with uncertain provenance; hypothetical proteins are typically enriched in mobile genetic elements); (iii) plasmid/viral proteins (sequences that have closest homologs in cellular organisms but are annotated as typical viral or plasmid proteins, such as structural components of virions, diverse methyltransferases, integrases, invertases, transposases and so on); (iv) viral proteins (encoded in genuine viral genomes).