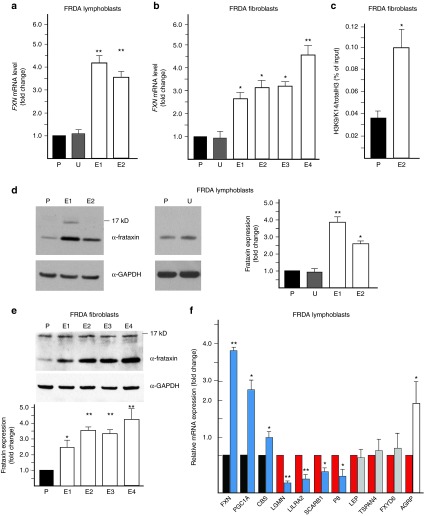
Figure 4.
Excision of the GAA tract increases frataxin expression and corrects the molecular phenotype of FRDA cells. (a) Determination of FXN mRNA expression using qRT-PCR in the ZFN-edited FRDA lymphoblast clones (E1 and E2) relative to the non-GAA edited control (P) and untransfected control (U). (b) Determination of FXN mRNA expression using qRT-PCR in the ZFN-edited FRDA fibroblast clones (E1–E4) relative to the non-GAA edited control (P) and untransfected control (U). (c) Chromatin immunoprecipitation with an antibody specific to acetylated lysines 9 and 14 on histone H3 (H3K9K14ac) in ZFN-corrected FRDA fibroblast clone E4 and non-GAA edited FRDA fibroblasts (P). (d) Analysis of frataxin expression in ZFN-edited FRDA lymphoblast clones E1 and E2 as determined by western blot. P designates non-GAA edited FRDA lymphoblasts; U designates untransfected FRDA lymphoblasts. Quantitative analysis of frataxin expression is shown in the graph. (e) Analysis of frataxin expression by western blot in the ZFN-corrected FRDA fibroblast clones (E1–E4) relative to the non-GAA edited control (P). Quantitation of the western blot is shown below. (f) The effect of ZFN correction on the FRDA signature of lymphoblast cells. A set of FRDA expression biomarkers37 was analyzed by qRT-PCR. The expression of the selected mRNAs was normalized to the non-GAA edited FRDA lymphoblasts. The mRNAs known to be underexpressed in FRDA lymphocytes are indicated as black bars while the overexpressed are shown as red bars.37 Editing of the expanded FXN allele reverses the changes related to frataxin deficiency in six biomarkers (blue bars), does not affect the expression of three mRNAs (gray bars), and has an inverse effect on expression of a single mRNA (white bar). The designations * and ** indicate statistically significant differences with P-values <0.05 and <0.01, respectively. The error bars represent SD of three or more experiments.