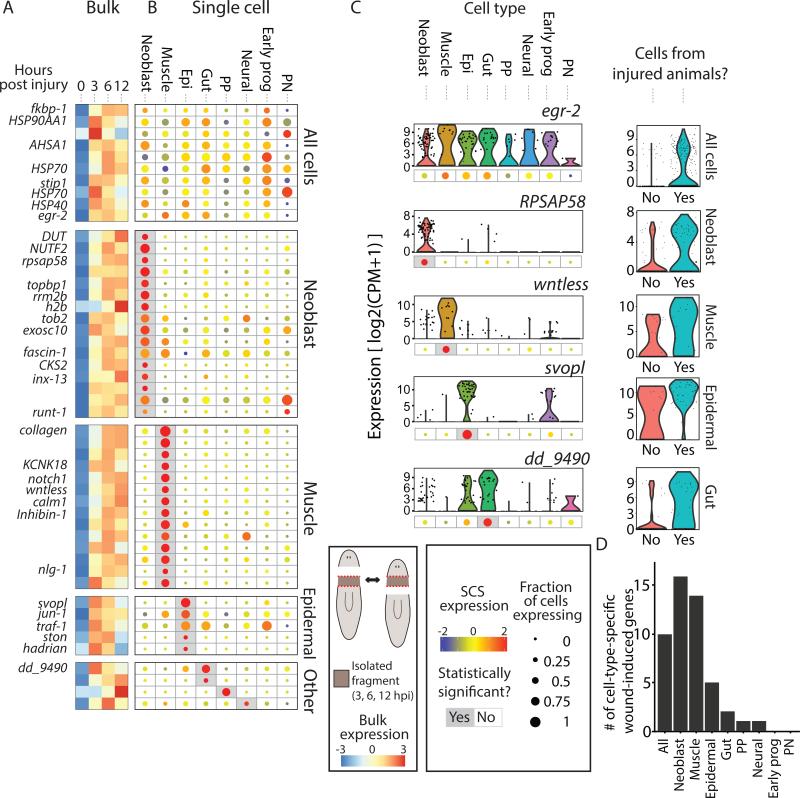
Figure 2. Cell-type-specific expression of wound-induced genes.
(A) The expression of wound-induced genes, as detected by bulk RNA-sequencing, is shown at different time points (0, 3, 6, and 12 hpi). Shown is the average expression of the anterior- and posterior-facing time courses. Rows and columns represent genes and time points, respectively. Gene expression is colored according to the z-transformed expression (z-score range is −3 to 3). Shown are wound-induced genes for which cell-type specificity was determined. (B) The corresponding cell-type-specific gene expression is shown in a dot-plot map. Dot size represents the proportion of cells expressing the gene (see key; 0-1), and the color represents normalized expression in cells expressing the gene (blue-to-red, low-to-high expression). Gray background represents statistically significant enrichment in a cell type (FDR ≤ 0.01; Extended experimental procedures). Genes are ordered according to their controlled enrichment p-value. Genes assigned to the All cells were overexpressed following wounding in multiple cell types (Methods). Cell-type acronym labels: NB, Neoblasts; Epi, epidermal lineage; Early prog, early epidermal progenitors; PP, para- pharyngeal; PN, protonephridia). (C) Left panels: Representative genes with wound-induced expression in different cell types. Expression across cell types is shown in violin plots with corresponding dot-plots beneath. Right panels: violin plots comparing the expression in cells of the cell type the gene was found to be enriched in between uninjured and injured animals. (D) Summary of the detected cell-type-specific wound-induced genes.