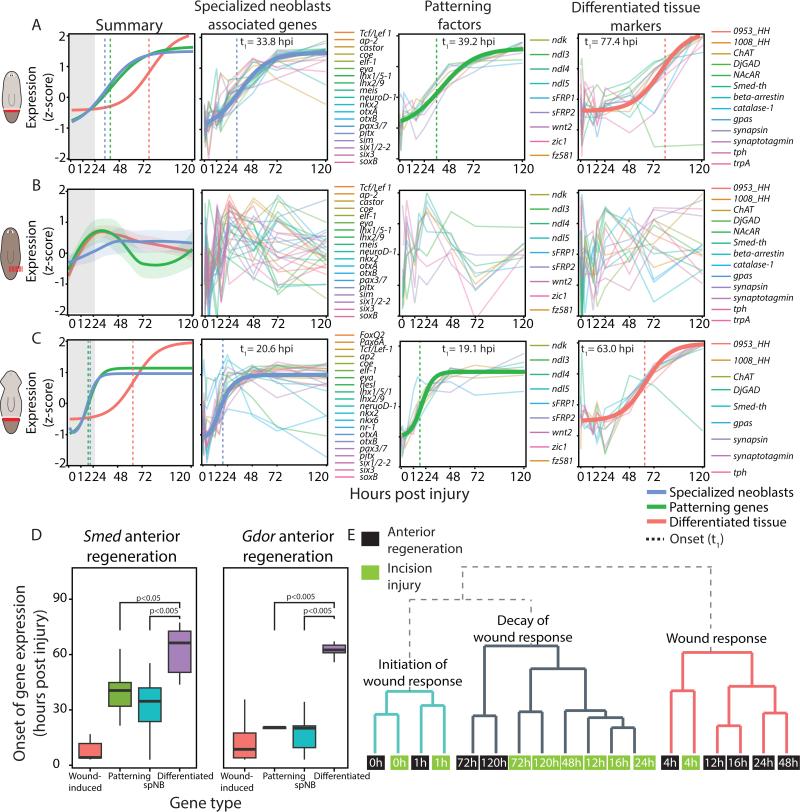
Figure 6. Injury-specific regeneration occurs in a temporally defined order.
(A) Summary panel: shown is a fit of the normalized median expression of neoblast specialization-associated genes, injury-specific patterning factors, and terminally differentiated tissue markers (blue, green and red, respectively). Matching colored vertical lines mark the onset times of the corresponding group of genes. Gray box highlights the wound-response phase. Other panels: bold lines represent impulse model fit of the genes used for modeling the dynamics of the group; thin lines represent individual genes. Onset time is marked by a vertical dashed line. (B) The genes used for panel A were plotted with the incision time-course data in which there was no missing tissue. Shown is a loess fit (bold lines) and confidence interval of the z-scores for each class of genes (lightly colored area) as the data could not be fit to the impulse model. Individual panels show a non-specific response following wounding. (C) A similar analysis performed on anteriorly regenerating G. dorotocephala revealed a similar order of events to amputation in S. mediterranea. (D) Box plot showing the onset time of different groups of genes following amputation. Boxes represent the interquartile range, thick lines are the median. Statistical significance was tested by a ks-test. (E) Dendrogram illustrating the similarity of gene expression of wound-induced genes in samples from the anterior regeneration and the incision time courses. Each node represents a sample (0-120 hpi; green and black nodes, incision and anterior samples, respectively). Annotations on the tree represent the interpretation of samples in clade.