Fig. 2.
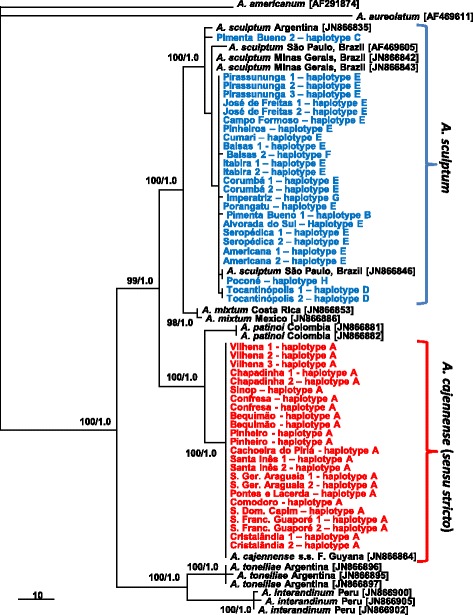
Bayesian inference phylogenetic tree with Maximum parsimony (MP) support values of the ITS2 rDNA sequences generated in the present study for the ticks Amblyomma cajennense (sensu lato), including 24 genotypes of A. cajennense (sensu stricto), indicated in red, and 26 genotypes of A. sculptum, indicated in blue. Each genotype is labelled as: Municipality name, number of the tick specimen, and haplotype code, as shown in Table 2. ITS2 sequences for Amblyomma americanum and A. aureolatum were used as the outgroup. Numbers at nodes are support values derived from bootstrap (1,000 replicates for MP/posterior probabilities for Bayesian inference analysis). Numbers in brackets are GenBank accession numbers
