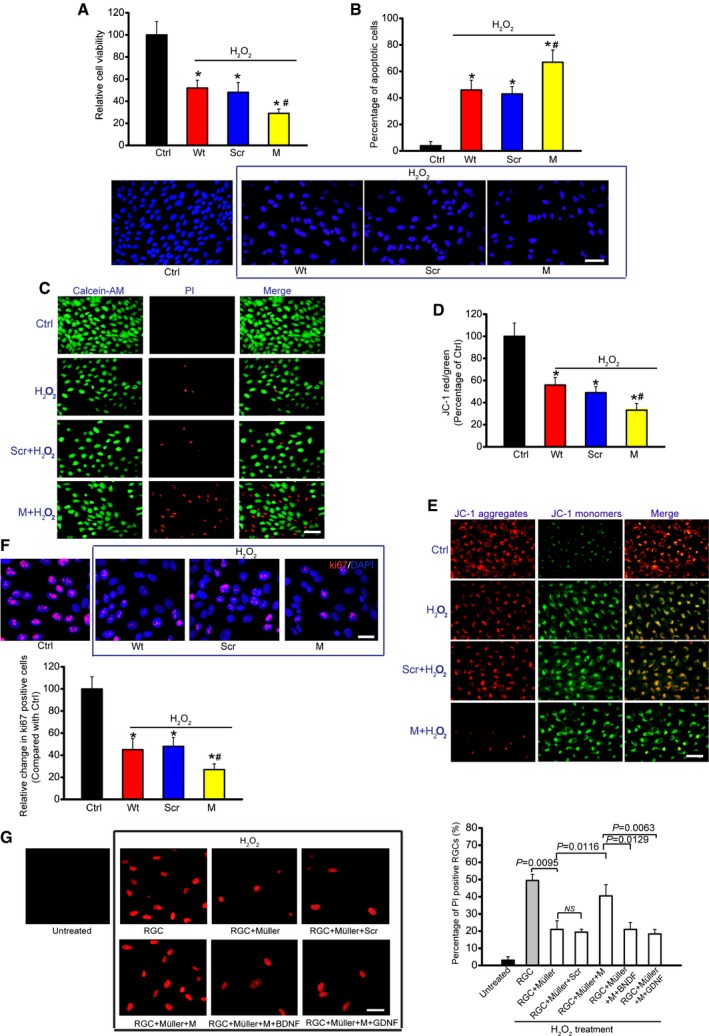
Primary RGCs were transfected with scrambled (Scr) shRNA, MALAT1 shRNA, or left untreated and then exposed to H
(50 μm) for 48 h. The group without any treatment was taken as the control (Ctrl) group.
-
A
Cell viability was determined using MTT method [n = 5 independent experiments; analyzed by two‐sided Student's t‐test; *P = 0.0285 (Wt), *P = 0.0246 (Scr), *P = 0.0132 (M), #
P = 0.0311].
-
B
Apoptotic cells were analyzed using Hoechst staining and quantitated [n = 5 independent experiments; analyzed by two‐sided Student's t‐test; *P = 0.0211 (Wt), *P = 0.0283 (Scr), *P = 0.0076 (M), #
P = 0.0289]. Scale bar, 50 μm.
-
C
Dead or dying cells were analyzed using calcein‐AM/PI staining from three independent experiments. Green, live cells; red, dead, or dying cell. Scale bar, 50 μm.
-
D, E
Primary RGCs were incubated with JC‐1 probe at 37°C for 30 min, centrifuged, washed, transferred to a 96‐well plate, and assayed using a fluorescence plate reader [n = 5 independent experiments; analyzed by two‐sided Student's t‐test; *P = 0.0212 (Wt), *P = 0.0299 (Scr), *P = 0.0147 (M), #
P = 0.0349], and observed using a fluorescence microscope. Red fluorescence, JC‐1 aggregates; green fluorescence, JC‐1 monomers (E). Scale bar, 50 μm.
-
F
Ki67 staining and quantitative analysis was performed to detect RGC proliferation [n = 5 independent experiments; analyzed by two‐sided Student's t‐test; *P = 0.0178 (Wt), *P = 0.0235 (Scr), *P = 0.0089 (M), #
P = 0.0186]. Scale bar, 20 μm. DAPI, blue; Ki67, red.
-
G
Primary RGCs were co‐cultured with Müller cells. Müller cells were transfected with MALAT1 (M) siRNA or scrambled (Scr) siRNA and then treated with or without BDNF (1 μg/μl) or GDNF (1 μg/μl). After these treatments, the experimental groups were exposed to H2O2 (50 μm) for 48 h. PI staining and quantitative analysis was performed to detect the dead or dying RGCs (n = 5 independent experiments; analyzed by one‐way ANOVA with Bonferroni post hoc). Scale bar, 20 μm.
Data information: * indicates significant difference compared with Ctrl group.
indicates significant difference between the marked groups. Data are represented as mean ± SEM.