Fig. 2.
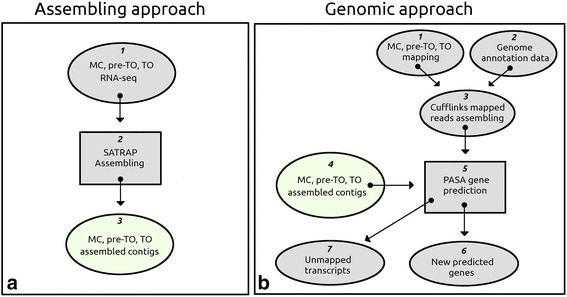
Genome annotation enrichment. a (1) the RNA-seq data referred to MC, pre-TO and TO were cleaned for the presence of contaminants (Ribosomal and bacterial sequences) and then (2) assembled and color translated using the program SATRAP. The resulted assemblies (3) contain also the transcripts that did not map in the reference genome because of the lacking of genome information. b the mapping information of MC, pre-TO and TO phases (1) as well as the genome annotation data (2) were passed to the program CUFFLINKS (3). The parsimonious dataset of transcripts produced by the program CUFFLINKS and the assembling information of each considered developmental phase (4) (coming from Panel A step 3) were analyzed by the program PASA (5) to produce a new gene prediction consistent with the reference genome sequence (6). Unmapped contigs (7) were reconsidered for further analysis
