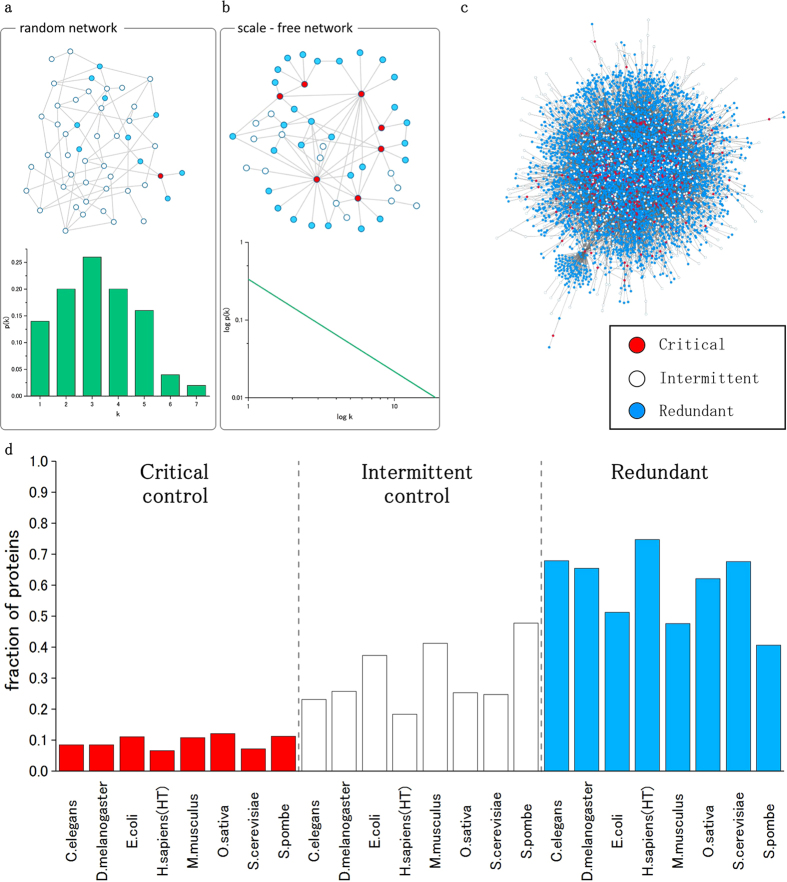
Figure 1.
A random network (a) and a scale-free network (b) and their corresponding degree distribution. The example shows that the network structure strongly affects the controllability roles of the nodes. Whereas in random networks the critical nodes are almost absent, in scale-free networks, they have a noticeable representation impacting the controllability of the network. Random networks, by contrast, are more flexible because they depend on intermittent control nodes. The proposed algorithmic procedure relies on the structure of the scale-free networks and on the existence of highly connected nodes to pre-determine a large number of critical and redundant nodes without using ILP. (c) The results of the algorithm computed on the human protein interaction network. (d) The fraction of proteins for each control category in all analysed PPI networks. The set of proteins involved in critical control is very small (less than 10% in almost all cases). The smallest critical control set is found in the H. sapiens and represents 6% of all proteins.