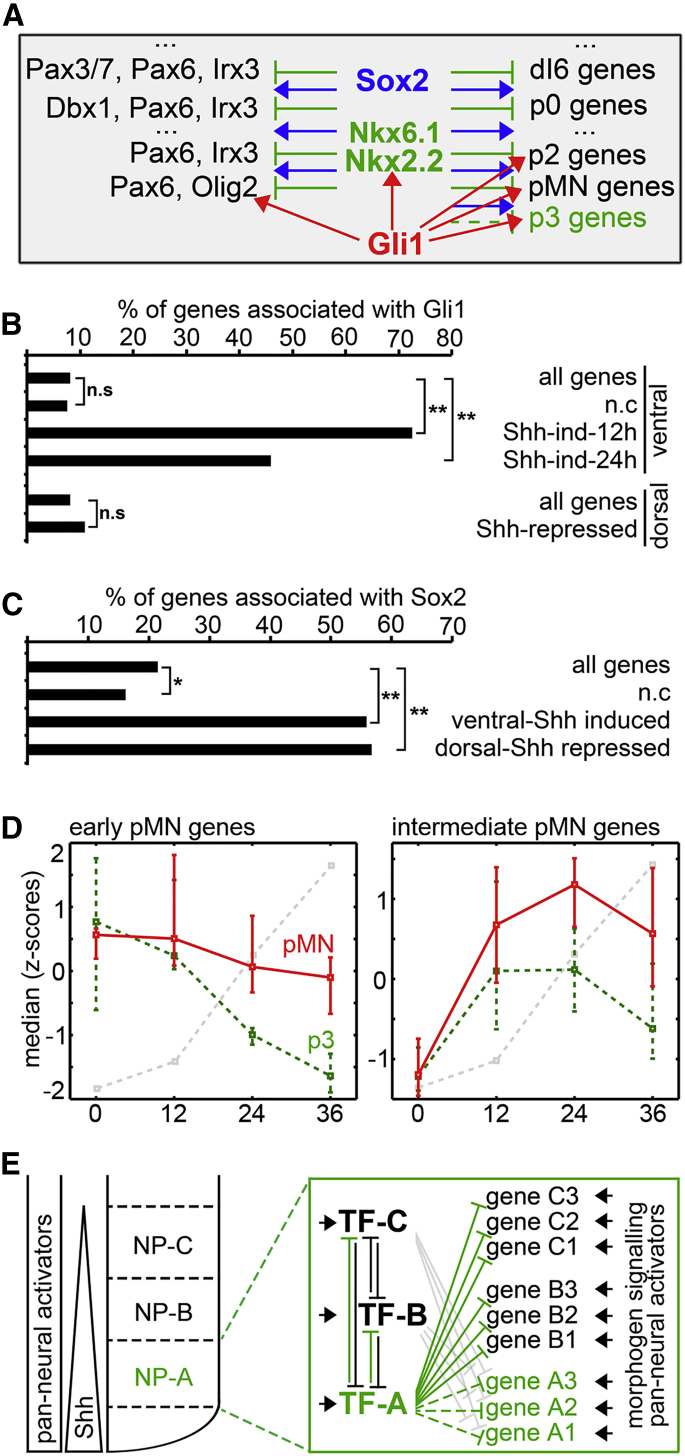
Figure 6.
Shh via Gli and Sox2 Provide Direct Positive Input Into the Expression of Ventral Genes
(A) Experimental rationale: to test whether Shh directly provided the positive input into the transcription of ventral genes, Gli1 binding (Peterson et al., 2012) was analyzed with respect to genes induced and repressed by Shh signaling. To test whether the pan-neural TF Sox2 provided direct input into transcription of neural progenitor genes, Sox2 binding was analyzed with respect to genes expressed in ventral and dorsal domains.
(B) 73% of the genes upregulated by Shh at 12 hr and 46% of those upregulated at 24 hr in ventral progenitors (Figure S4A and Table S1, sheet S4) are associated with binding of Gli1. By contrast, few of the dorsal genes, repressed by Shh signaling, in p3 and/or pMN are associated with Gli1 binding. ∗∗p(χ2) < 0.001, n.s, non-significant.
(C) Sox2 binds close to the majority of both ventral (induced by Shh in p3 or pMN at 12 or 24 hr) and dorsal (repressed by Shh in p3 or pMN at 24 hr) genes. ∗∗p(χ2) < 0.001, ∗p(χ2) < 0.005. See also Figures S4B and S4C.
(D) Median expression levels of early and intermediate pMN genes (genes expressed higher in pMN than p3, see Figure 2D) in pMN cells (red line) and p3 cells (green dotted line). The lines represent the median values of the indicated groups of genes and the error bars correspond to the 10th and 90th percentile values of each group. Note that the pMN genes are induced in both pMN and p3 cells simultaneously and they are only repressed in p3 later, at the time Nkx2.2 (gray dotted line) is activated at 24 hr. This suggests the common activatory inputs in p3 and pMN domains. 56% of intermediate genes are induced by Shh and 67% of them are bound by Gli1.
(E) Model summary. Four design features of the neural tube GRN. First, broadly expressed and promiscuous activating inputs from morphogen mediators and other transcriptional activators promote the transcriptional programs of multiple progenitor domains (A, B, C). Second, specific cell identity is determined by a network of transcriptional repressors (TF-A, TF-B, TF-C), these ensure cells select a single identity by repressing all inappropriate cell fates. Third, the repressors directly inhibit the expression of not only other repressors, but all the “effector” genes specific for other progenitor domains. This counteracts the direct positive inputs into all genes. Finally, the regulatory input into target genes is combinatorial and it is the integration of multiple, sometimes conflicting, inputs that determines cell identity.