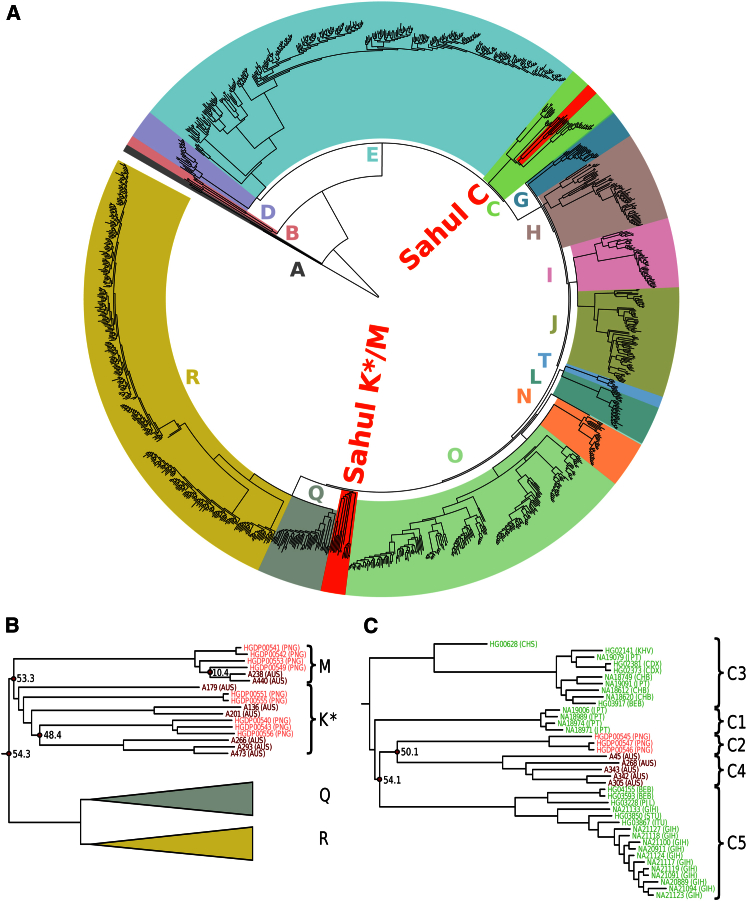
Figure 1.
Phylogenetic History of Aboriginal Australian Y Chromosomes
(A) A maximum-likelihood phylogeny was inferred from the Y chromosome data of 1,269 males from worldwide populations, including Aboriginal Australians, using RAxML [14]. High-level haplogroups are colored and labeled along the tree. The two clades that contain the Y chromosomes indigenous to the continent of Sahul (from the Aboriginal Australian and Papuan samples) are indicated in bright red.
(B) The phylogeny of Y chromosomes in haplogroups K∗ and M. This detailed view of a part of the larger tree displayed in (A) focuses on chromosomes in haplogroups K∗ and M. Haplogroups Q and R, which are the closest relatives to K∗ and M in the phylogeny, are represented schematically because they contain very large numbers of samples. Aboriginal Australian and Papuan samples are colored in two different shades of red for easier visual separation. Sample names and population origins are displayed at branch tips (AUS, Aboriginal Australian; PNG, Papua New Guinean). Divergence times in units of thousands of years are indicated on key nodes that correspond to divergences between groups of samples from different populations or haplogroups.
(C) The phylogeny of Y chromosomes in haplogroup C. Sample names and population origins are displayed at branch tips (AUS, Aboriginal Australian; PNG, Papua New Guinean; CHS, Southern Han Chinese in China; KHV, Kinh in Ho Chi Minh City, Vietnam; JPT, Japanese in Tokyo, Japan; CDX, Chinese Dai in Xishuangbanna, China; CHB, Han Chinese in Bejing, China; BEB, Bengali in Bangladesh; PJL, Punjabi in Lahore, Pakistan; GIH, Gujarati Indian in Houston, Texas; STU, Sri Lankan Tamil in the UK; ITU, Indian Telugu in the UK). We note that due to factors associated with missing data arising from the low sequencing coverage of the 1000 Genomes samples, the branch lengths displayed here are not strictly proportional to time.
See also Table S1.