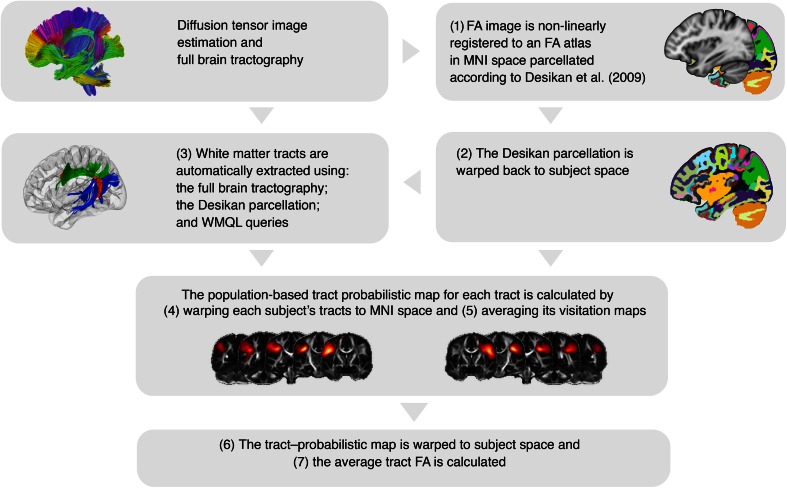
Fig. 1.
DTI processing steps. The input to the pipeline include diffusion tensor imaging (DTI) data, a fractional anisotropy (FA) template with a Desikan et al. (2006) parcellation superimposed, and a set of queries in White Matter Query Language (WMQL) describing the sections of the superior longitudinal fasciculus (SLF) to be obtained. The pipeline involves an initial step for each subject estimating the DTI image and calculating from this image the FA map and a full-brain tractography. Next, child-specific probabilistic template maps are computed using the following procedures: (1) the FA image is registered to the Montreal Neurological Institute (MNI) template and (2) the Desikan parcellation is warped back to the subject using the inverse transformation map. Then, (3) the WMQL and the Desikan parcellation are used to obtain the sections of the SLF for each subject from the full-brain tractography, and (4) these tracts are warped to the template space to (5) generate the population-based tract probabilistic maps by averaging the visitation maps of all subjects. Finally, (6) these probabilistic maps are warped back to subject space and (7) used to compute the average FA of each tract for each subject and perform statistical analysis