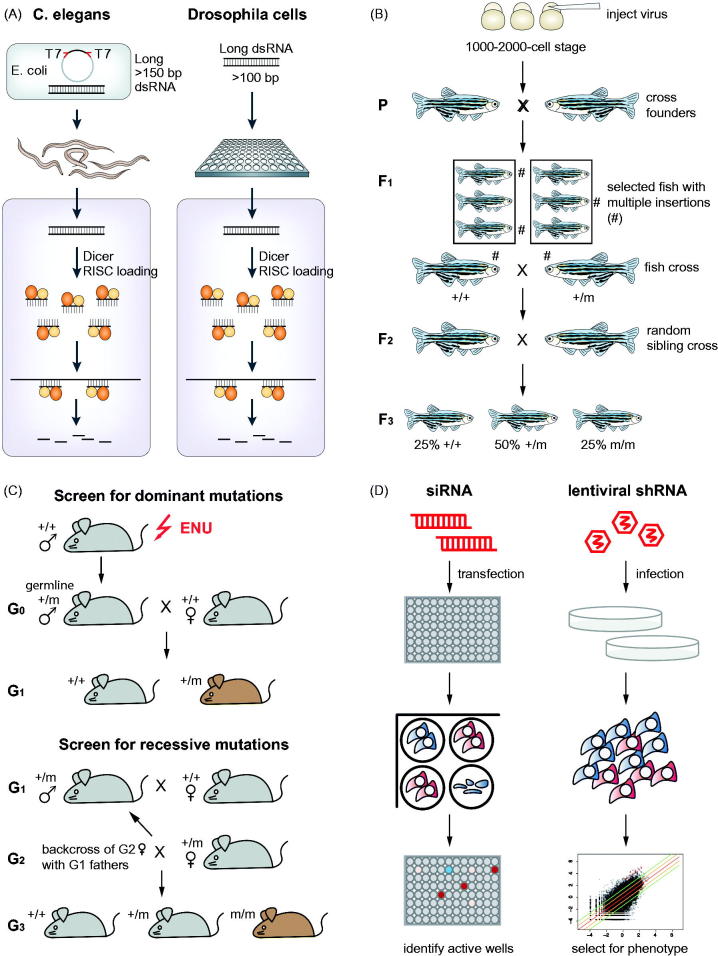
Figure 2.
Screening strategies in different model organisms. (A) Schematic overview of RNAi screening approaches in C. elegans by ingestion of E.coli and in Drosophila cells by bathing (modified from Boutros & Ahringer (2008)). Long double-stranded RNAs are introduced into the respective organisms and diced intracellularly into small-interfering RNAs (siRNAs). This results in many different siRNAs targeting a single transcript. (B) Outline of insertional mutagenesis screen in zebrafish (adapted by permission from Macmillan Publishers Ltd.: Nature Reviews Genetics (Patton & Zon, 2001), (c) 2002). Embryos are injected with a retrovirus at a 1000–2000-cell stage. These embryos are raised (= founder generation P), mated and the mutations transmitted to the F1 generation. Individual fish from the F1 generation with multiple insertions are selected and further crossed with each other to generate the F2 generation. Siblings of each F2 family are crossed with each other to generate homozygous mutations. (C) Overview of screening approaches using ENU induced mutagenesis in mouse. For dominant mutations, ENU mutagenized males carrying mutations in their germ lines are crossed with wild-type females (G0). Dominant mutations will be detected in the G1 generation. For recessive mutations, males of the G1 generation carrying mutations are crossed with wild type females. Then females of the resulting G2 generation are backcrossed with G1 males, thereby generating the G3 generation that potentially carries individuals with homozygous mutations. (D) Arrayed versus pooled loss-of-function screens in cancer cell lines. siRNAs are used for arrayed screens in a multi-well plates. Each well harbors a distinct gene knockdown. Candidate genes are detected by measuring signal levels (e.g. luminescence) of individual wells. In contrast, pooled loss-of-function screens rely on viral infection of cells with shRNA vectors. Each vector contains a barcode allowing identification of the specific shRNA. Pools of cells with different gene knockdowns are generated and cultured for several doubling times. Depletion of cells with specific gene knockdowns is detected by sequencing of barcodes and measuring their relative abundance at different time points.