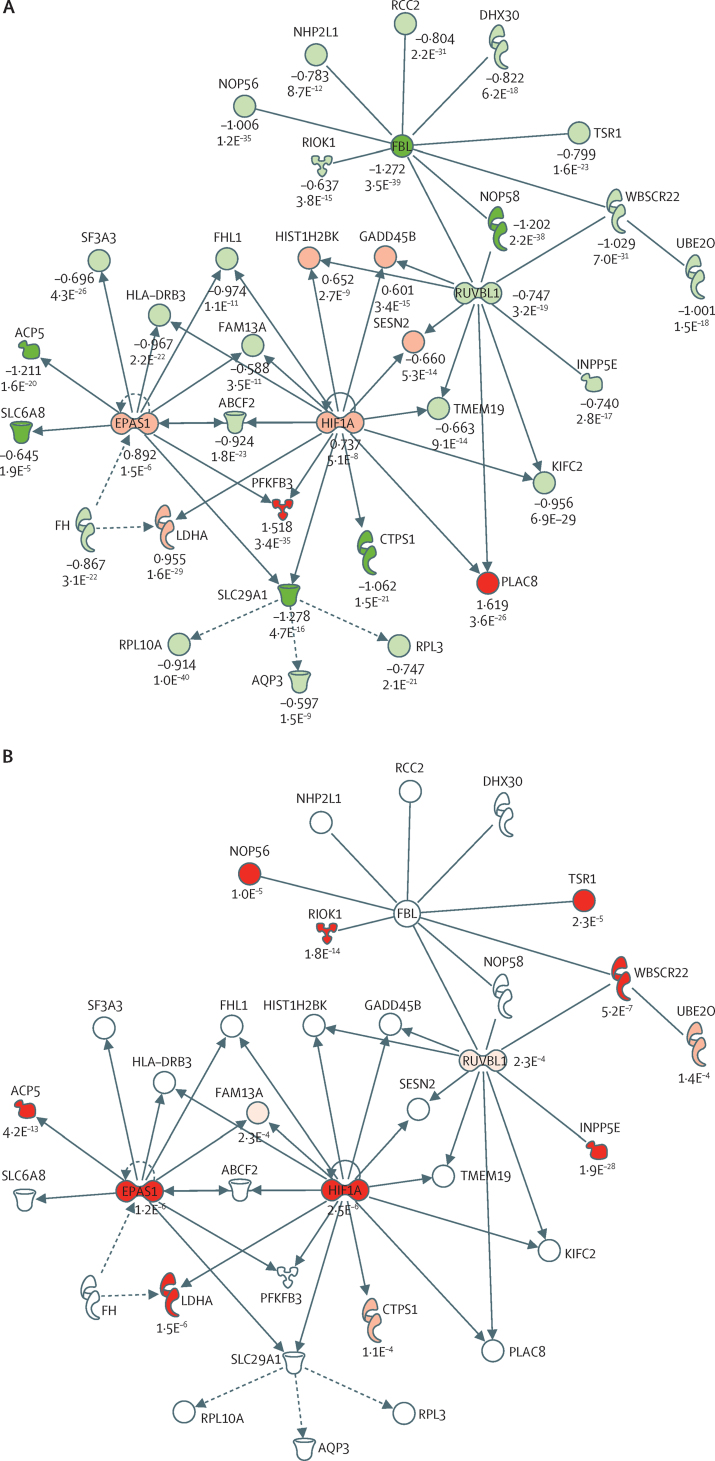
Figure 3.
Differentially expressed networks between SRS
The network with the lowest p value (p=1 × 10−35) identified from differential expression analysis of SRS1 versus SRS2 in the discovery cohort included 35 genes, with HIF1A and EPAS1 as nodes (A). Log FC shown with FDR. Sepsis cis-eQTL affecting genes in hypoxia network (B). Presence of cis-eQTL shown by red molecules with p values. eQTL=expression quantitative trait loci. SRS=sepsis response signature. FC=fold change. FDR=false discovery rate.