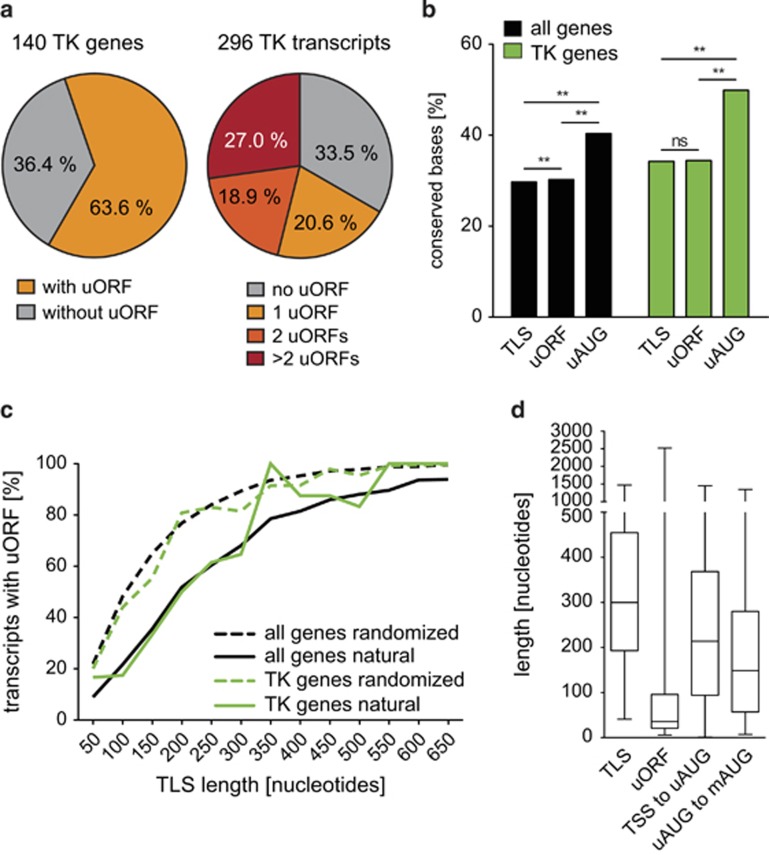
Figure 1.
Presence, conservation and position of uORFs in human tyrosine kinases and all human genes. (a) Pie charts indicating the prevalence of uORFs in TK genes and the number of uORFs per transcript for alternative TK mRNAs. (b) Bar graph displaying the frequency of conserved bases within TLSs, uORFs and uAUGs of TKs and all human genes. Note that columns labeled ‘TLS' do not include uORF bases and columns labeled ‘uORF' do not include uAUGs, respectively. For genome-wide transcript analyses, Hg19 annotations were downloaded from the UCSC genome browser database.15 Of 44 113 entries, we excluded 7794 non-coding RNAs, 1861 sequences not aligning to autosomes or sexual chromosomes, and 1237 transcripts lacking a TLS. Finally, we positively selected the 32 332 mRNAs with unique genome alignments corresponding to 17 893 distinct genes (ftp://ftp.ncbi.nih.gov/refseq/H_sapiens/mRNA_Prot/). Conservation was defined by mapping the respective genomic positions to the ‘100 vertebrates conserved elements' provided by UCSC (http://genome.ucsc.edu/cgi-bin/hgTrackUi?g=cons100way). Statistical significance was analyzed using the χ2 test and indicated as **P<0.01 and NS (not significant). (c) Graph indicating the prevalence of uORFs in respect to the length of the TLS for TKs and all human genes (solid green and black lines, respectively). Dashed lines indicate expected uORF frequencies for TKs and all human genes following a 10 000-times randomization of TLS nucleotides (green and black, respectively). (d) Box plots summarizing the variable length of TLSs and uORFs in TK transcripts. The graph indicates the range of nucleotide distances from the transcriptional start site (TSS) to the uAUG and from the uAUG to the main AUG (mAUG).