Figure 4. Characterisation of behavioural patterns for different H. pylori mutants in genes coding for TlpD-interacting proteins (swim assay).
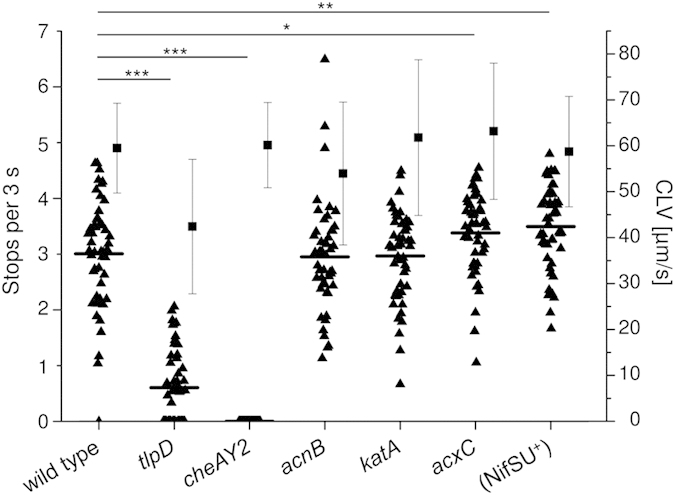
Behavioural patterns of H. pylori were determined by tracking assays using 50 bacteria per strain under previously determined steady state conditions of low bacterial energy yield13,26 in RPMI 1640 + 3% FCS at 37 °C and ambient air enriched with 5% CO2. Bacteria were collected from blood agar plates after 20 h of growth and resuspended in the liquid medium of low energy yield. Under these conditions, stopping frequency is dominated by TlpD26. Stops per three seconds (black triangles with mean) and curvilinear velocity (CLV; average with standard deviation as black squares with whiskers) were quantified for the different mutants. Significance values for differences between the strains were calculated by Student’s t test.
