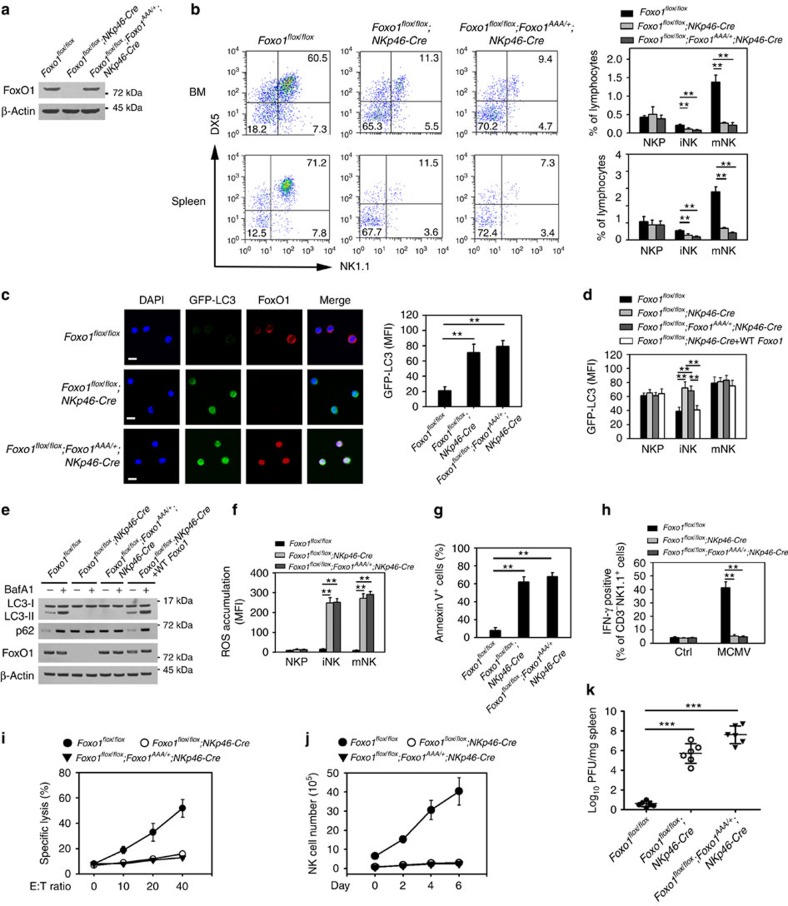
Figure 7. FoxO1 deficiency or Foxo1AAA mutant abolishes autophagy induction and NK cell development.
(a) FoxO1 of BM NK cells (CD3−NK1.1+) was deleted in Foxoflox/flox;NKp46-Cre mice and Foxo1flox/flox;Foxo1AAA/+;NKp46-Cre mice by immunoblotting assay. (b) NK cell development is impaired in Foxo1-deficient or Foxo1AAA-mutant mice. Propidium iodide (PI)- negative BM and spleen NK cells in Foxo1flox/flox, Foxo1flox/flox;NKp46-Cre and Foxo1flox/flox;Foxo1AAA/+;NKp46-Cre mice were analysed by flow cytometry. For each group, n=7 mice. (c) iNK cells were separated from the BM of GFP-LC3;Foxo1flox/flox, GFP-LC3;Foxo1flox/flox;NKp46-Cre and GFP-LC3;Foxo1flox/flox;Foxo1AAA/+;NKp46-Cre mice for immunostaining. GFP-LC3 MFI of each cell was analysed and show as means±s.d. **P<0.01. Scale bar, 5 μm. (d,e) For WT FoxO1 rescue, WT NKPs from GFP-LC3 mice were adoptively transferred into FoxO1flox/flox;NKp46-Cre;GFP-LC3 mice. After three weeks, autophagic flux of NK cells in the indicated mice expressing GFP-LC3 were analysed by flow cytometry (d). iNKs from the indicated mice were isolated and treated with BafA1 followed by western blotting (e). (f) ROS were detected in different NK cells from Foxo1flox/flox, Foxo1flox/flox;NKp46-Cre and Foxo1flox/flox;Foxo1AAA/+;NKp46-Cre mice. (g) Apoptosis of BM CD3−NK1.1+ NK cells from the indicated mice were stained with Annevin V. (h) The indicated mice were infected with MCMV (1 × 105 PFU) for 3 days and intracellular IFN-γ of CD3−NK1.1+ splenic NK cells were analysed by flow cytometry. (i) Foxoflox/flox, Foxo1flox/flox;NKp46-Cre and Foxo1flox/flox;Foxo1AAA/+;NKp46-Cre mice were infected with MCMV for 3 days. NK cells were separated from spleens and incubated with 51Cr-labelled Yac1 cells for cytotoxicity assay. Specific lysis of Yac1 cells was analysed by 51Cr release, and shown as means±s.d. (j) The indicated mice were infected with MCMV for the indicated days, and NK (CD3−NK1.1+) cell number was analysed by flow cytometry. (k) Virus titres from MCMV-infected Foxoflox/flox, Foxo1flox/flox;NKp46-Cre and Foxo1flox/flox;Foxo1AAA/+;NKp46-Cre mice were analyzed after 3 days infection (n=6 mice). All data represent at least three independent experiments and calculated data are shown as means±s.d. **P<0.01; ***P<0.001. For b–d,f,g and k, a two-way analysis of variance post hoc Bonferroni test was used.