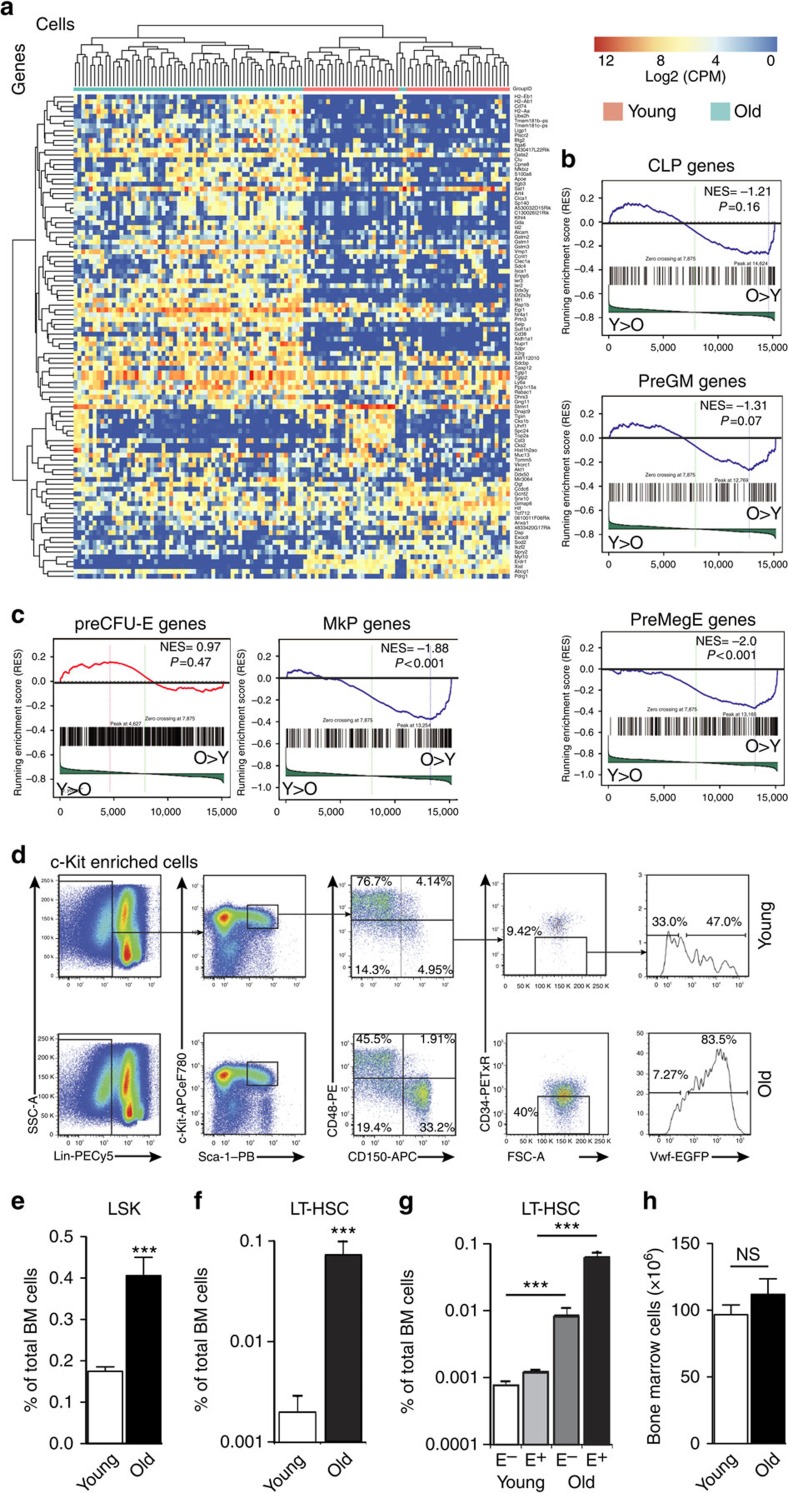
Figure 1. Expansion of platelet-primed HSCs in old mice.
(a) Hierarchical clustering of young and old LSKCD150+CD48− HSC single-cell transcriptomes using the top 100 variable genes from components 1–3 of combined PCA of young and old HSCs. Data are from two independent experiments (young mice N=1; old mice N=1). (b) Signature enrichment plots from GSEA analysis using CLP, preGM and preMegE signature gene sets. The values indicated on individual plots are the Normalised Enrichment Score (NES) and P-value of the enrichment (permutation analysis). Note the significant upregulation of preGM, and preMegE signatures in old HSCs. (c) Signature enrichment as in b using CFU-E and MkP signature gene sets. Note the significant upregulation of the MkP signature in old HSCs. (d) Representative FACS plots for isolation of Vwf- and Vwf+ LT-HSCs (defined as LSKCD48−CD150+CD34− cells) in BM from young and old Vwf-EGFP transgenic mice after performing c-Kit enrichment. Numbers indicate percentage of the parental gate. (e) Frequencies of LSK cells in total BM in young (N=24) and old (N=5) mice. Data are from five independent experiments. Values show mean±s.e.m. ***P<0.001. (Student's t-test). (f) Frequencies of LSKCD48−CD150+CD34− LT-HSCs as percentage of live singlets in young (N=31) and old (N=7) mice. Note: Log10 scale. Data are from six independent experiments. (g) Frequencies of Vwf- and Vwf+ LT-HSCs as percentage of live singlets in young (N=31) and old (N=7) mice. E−, Vwf-EGFP−; E+, Vwf-EGFP+. Note: Log10 scale. Data are from six independent experiments. (h) Bone marrow cellularity of young (N=29) and old (N=7) Vwf-EGFP transgenic mice. Cellularity was determined from two iliac cristae, two tibiae and two femora after RBC lysis. Values show mean±s.e.m. ***P<0.001. (Student's t-test); NS, not significant. Data are from six independent experiments.