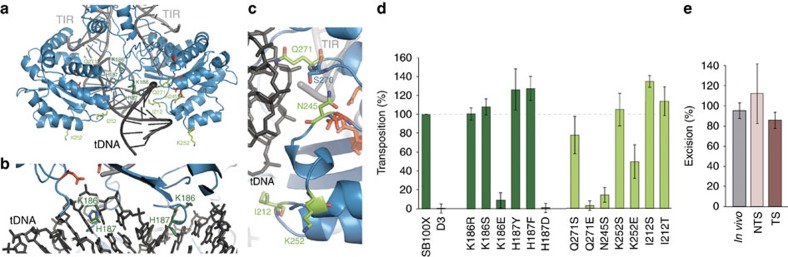
Figure 4. Structure-based mutagenesis maps tDNA binding and generates novel hyperactive transposases.
(a) TCC model with predicted tDNA-binding residues selected for mutagenesis. (b,c) Zoom-ups on residues predicted to be involved in tDNA recognition or bending (dark green, b) and residues lining the target-binding groove (light green, c). (d) Transposition assay for the mutants in HeLa cells. P values (based on one-sample t-test): K186R, 0.93; K186S, 0.22; K186E, 0.0019; H187Y, 0.069; H187D, 6.9 × 10−6; Q271S, 0.21; Q271E, 7.5 × 10−5; N245S, 0.0022; K252S, 0.68; K252E, 0.042; I212S, 0.0047; and I212T, 0.27. D3 indicates the negative control with the inactive E279D transposase. (e) Excision assays with the hyperactive I212S transposase in vivo and in vitro (monitoring cleavage on both DNA strands: NTS, TS). Results are normalized to SB100X. All error bars represent the s.e.m. for three independent experiments.