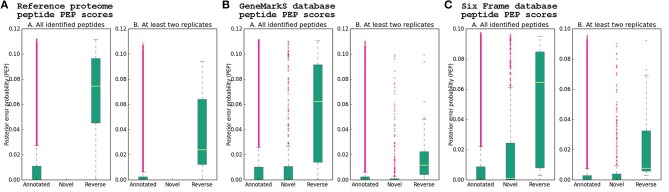
Figure 8.
Peptide PEP score distribution. The posterior error probability (PEP) score distribution of novel, annotated and reverse hit peptide identifications obtained using three different database searches are shown, using the PEP score of the best PSM obtained for every peptide, from the MaxQuant peptides.txt output file. (A) Reference proteome, (B) GeneMarkS database, (C) Six frame database. The whiskers represent 1.5 times the interquartile range (IQR) below and above the first and third quartile, respectively. Differences in the means of the groups from each database search were evaluated using the non-parametric Kruskal–Wallis test, which indicated significant differences between the means of novel and reverse peptide PEP scores in the GeneMarkS (adjusted p-value 5.50e-17) and six frame database (adjusted p-value 9.84e-13). The p-values assigned to the comparisons between annotated and novel groups for the GeneMarkS (adjusted p-value 8.75e-01) and six frame (adjusted p-value 1.89e-06) databases were much larger than the p-values assigned to the comparisons between novel and reverse groups. Significant differences were found in the annotated-reverse comparisons for all three databases—GeneMarkS (adjusted p-value 6.68e-21), six frame database (adjusted p-value 3.67e-19) and Reference proteome (adjusted p-value1.050e-31). The effect of excluding peptides seen only in a single replicate on the PEP score distributions is also apparent. The adjusted p-value assigned to the comparison between all six frame novel and annotated peptides (adjusted p-value 1.89e-06) was much smaller than the adjusted p-value assigned to the same comparison after excluding peptides seen only in a single replicate (adjusted p-value 1.77e-02). In contrast, the comparison between six frame novel and reverse peptides seen in at least two replicates was assigned an adjusted p-value of 2.87e-06, which is closer to the p-value assigned to the comparison between annotated and reverse peptides seen in at least two replicates (adjusted p-value 6.09e-08; see Supplementary Table 10 and Supplementary Data Sheet 5 for the results of Kruskal–Wallis tests and post-hoc pairwise comparisons).