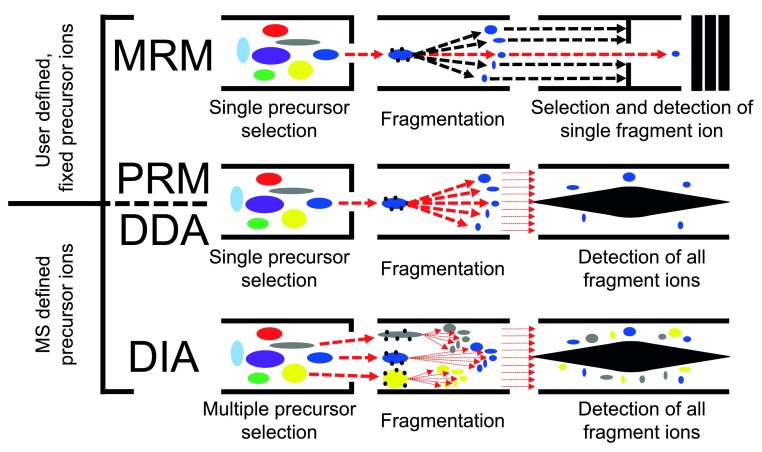
Figure 1. A cartoon schematic of how peptides are isolated, fragmented, and analyzed by a mass spectrometer working in data-dependent acquisition (DDA), multiple reaction monitoring (MRM), parallel reaction monitoring (PRM), or data-independent acquisition (DIA) modes.
In DDA, MRM, and PRM, single precursor ions are isolated, fragmented, and analyzed in an MS2 scan by the mass spectrometer. In DDA mode, the precursor ions are chosen by the instrument on the basis of abundance. In MRM and PRM, the precursor ions to be analyzed are fixed by the user. DIA is different form the methods above in that all precursor ions within a selected mass range are isolated, fragmented, and analyzed in a single MS2 scan. MS1, scan in which the peptide ions entering the mass spectrometer at a given time are identified; MS2, scan in which the fragments of all (or some) of the peptides that are in the mass spectrometer at a given time are identified.