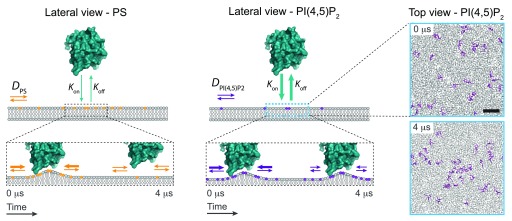
Figure 2. Schematic representation on how phosphoinositide (PI)-binding motifs can engage local demixing of PIs on cellular membranes.
As an example, lateral view of the ENTH domain of Epsin (PDB code 1H0A) in cyan upon binding to a membrane that contains monovalent lipids such as phosphatidylserine (PS) (in orange, left panel) or PI(4,5)P 2 (in magenta, right panel). Cyan arrows represent the K on/ K off rates of the ENTH domain binding on membranes, being faster for PS over PI(4,5)P 2. As a result, transient demixing of PI(4,5)P 2 molecules can take place. The diffusion of PS and PI(4,5)P 2 in the plane of the membrane is depicted by orange and magenta arrows, respectively. Right panel shows a top view of PI(4,5)P 2 clustering coarse-grain molecular dynamics simulations (as described in 19) on spontaneous membrane biding of an ENTH domain. The panels are snapshots at t = 0 μs and 4 μs of the individual position of PI(4,5)P 2 molecules (in magenta) along the simulation. Scale bar, 1 nm.