Figure 2.
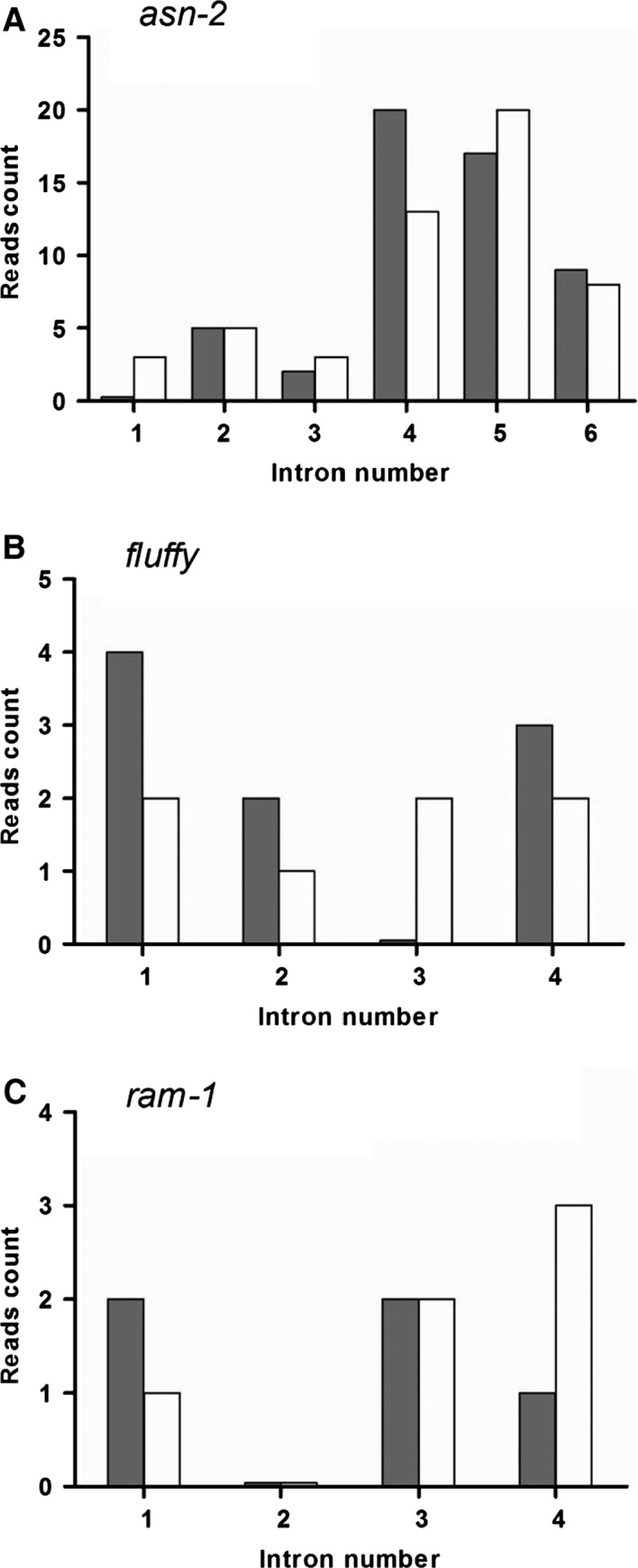
Intron retention evaluated by RNAseq in N. crassa. The Sequence Read Archive at the NCBI database (http://www.ncbi.nlm.nih.gov/sra) was used to quantify intron retention. Reads were then aligned to fasta files containing sequences of 50 nt representing each exon‐intron boundary for the genes of interest. The number of reads mapping at each boundary was used to estimate the prevalence of intron retention in each gene analyzed (5′‐ gray; 3′‐ white). (A) Asparagine synthetase 2 (asn‐2) (NCU04303); (B) C6‐zinc finger regulator (fluffy) (NCU08726); (C) farnesyltransferase (ram1) (NCU05999).
