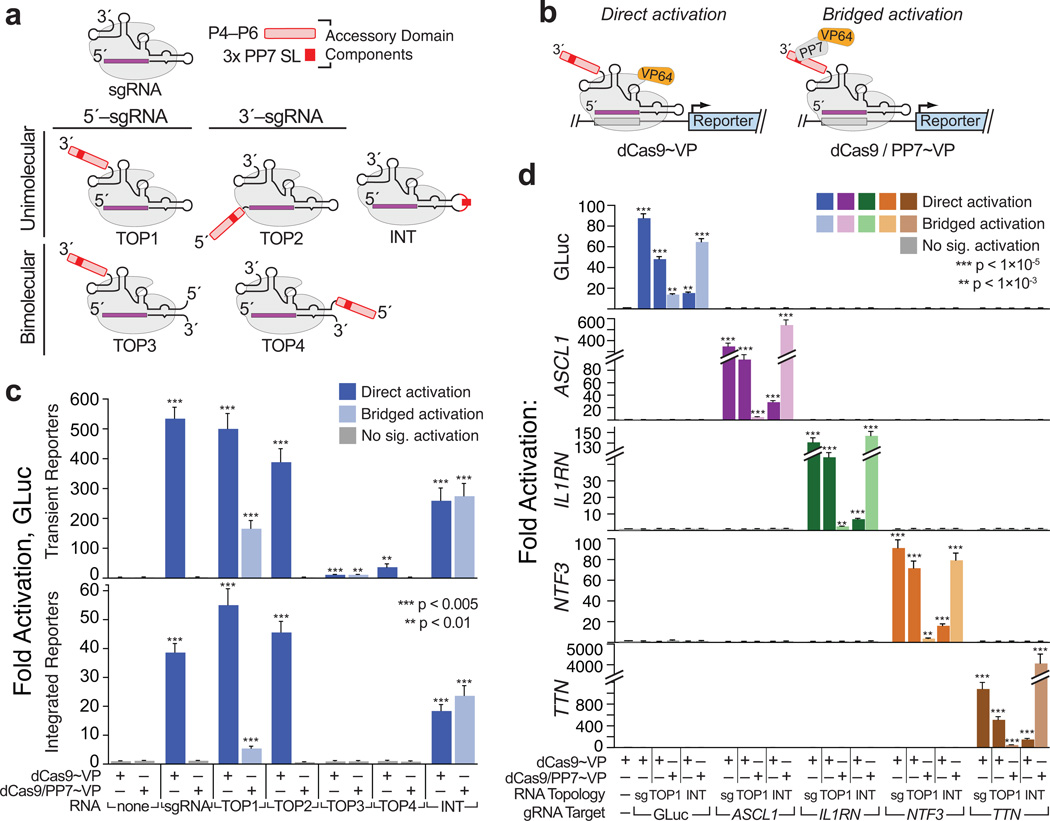
Figure 1. Large, structured RNA domains can be functionally appended onto the sgRNA scaffold at multiple points.
(a) Design of “TOP” topology constructs. Accessory RNA domains are detailed in (Supplementary Fig. 4a–b); expression constructs in (Supplementary Fig. 1c). (b) Schematics summarizing direct activation (left) and bridged activation (right) assays. (c) Luciferase reporter assays of the five topology constructs. (d) Targeting minimal (“sg”) and expanded (“TOP1” and “INT”) sgRNAs to endogenous loci. ASCL1, IL1RN, NTF3 and TTN. were each targeted using mixed pools of four gRNAs28, 29, 33 (Supplementary Table 2). GLuc activation was measured by luciferase assays; endogenous gene activation was measured using qRT-PCR (Supplementary Table 3). Values are means ± standard deviation, (n=3) Luciferase assays; (n=4) qRT-PCR. Student’s one-tailed t-test relative to cells expressing dCas9~VP alone.