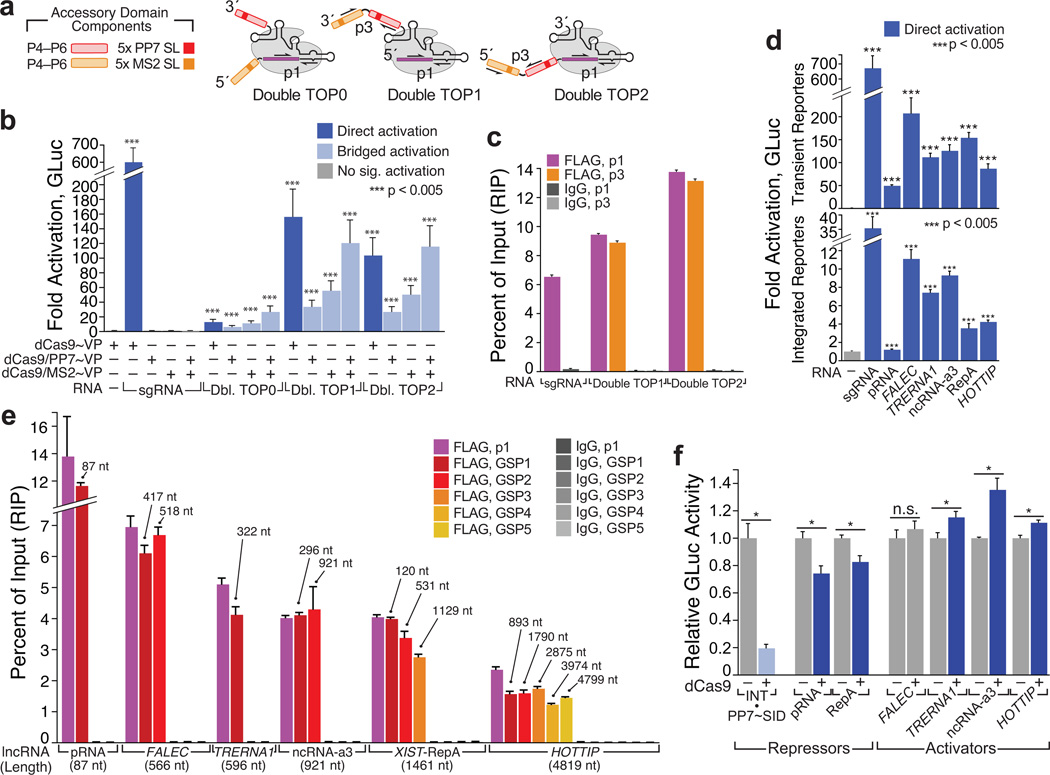
Figure 2. CRISPR-Display with artificial and natural lncRNAs.
(a) Design of “Double TOP” artificial lncRNA constructs. Accessory domains are detailed in (Supplementary Fig. 4a–d). The sgRNA core “p1” and domain-spanning “p3” qPCR primer pairs are indicated (Supplementary Table 3). (b) Direct and bridged activation assays using Double TOP (“Dbl TOP”) constructs, expressed from the CMV/3´Box backbone (Supplementary Fig. 6a). Transient reporter assays are shown. (c) RIP/qRT–PCR of dCas9•Double TOP1 and dCas9•Double TOP2. (d) sgRNAs appended with a battery of natural lncRNA domains (Supplementary Table 4) form functional complexes with dCas9~VP. Direct activation assays are shown. The minimal “pRNA” stem-loop34 was displayed internally; all other domains were appended on the sgRNA 3´ terminus. RNAs were expressed using the CMV/MASC system (Supplementary Fig. 6a).(e) LncRNA accessory domains remain intact in CRISP-Disp complexes. Immunopurified RNA was analyzed using qPCR primers targeting the sgRNA core (p1) and intervals along the each lncRNA domain (GSP1–GSP5, Supplementary Table 3). Above each primer set, the maximum distance from the sgRNA core is indicated. (f) Transient reporter assays with CRISP-Disp lncRNA constructs. Values are normalized relative to those of control cells expressing each lncRNA alone. For comparison, bridged repression with U6-driven INT, complexed with dCas9 and PP7~SID (Ref.39) is shown (left, light blue). Values are means ± standard deviation, (n=3) Luciferase assays; (n=4) qPCR. Student’s one-tailed t-test relative to cells expressing (b, d) dCas9~VP alone or (f) lncRNA alone. *, p < 0.05.