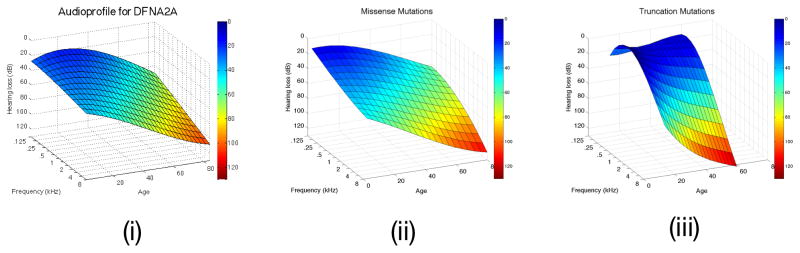
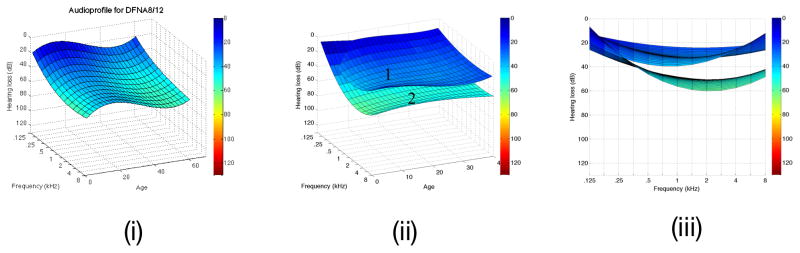
Figure 4.
Figure 4A. DFNA2A APSs is shown on the left (i). After clustering, two surfaces are identifiable, one generated from persons segregating missense variants of KNCQ4 (ii), while the other is based on truncating variants of KCNQ4 (iii). Note the more rapid high frequency rate of decline associated with truncating mutations in this gene. B. The DFNA8/12 APS is shown on the left (i). Two APS subclusters are generated by cluster analysis of this dataset (ii and iii, which displays data from the frequency perspectice). Multivariate analysis shows that these two subclusters do not reflect mutation type, mutation location, patient age or patient sex, suggesting that the observed differences may reflect secondary genetic modifiers. Table 1 shows that this hypothesis is reasonable given the known variability in the protein constituents of the tectorial membrane.