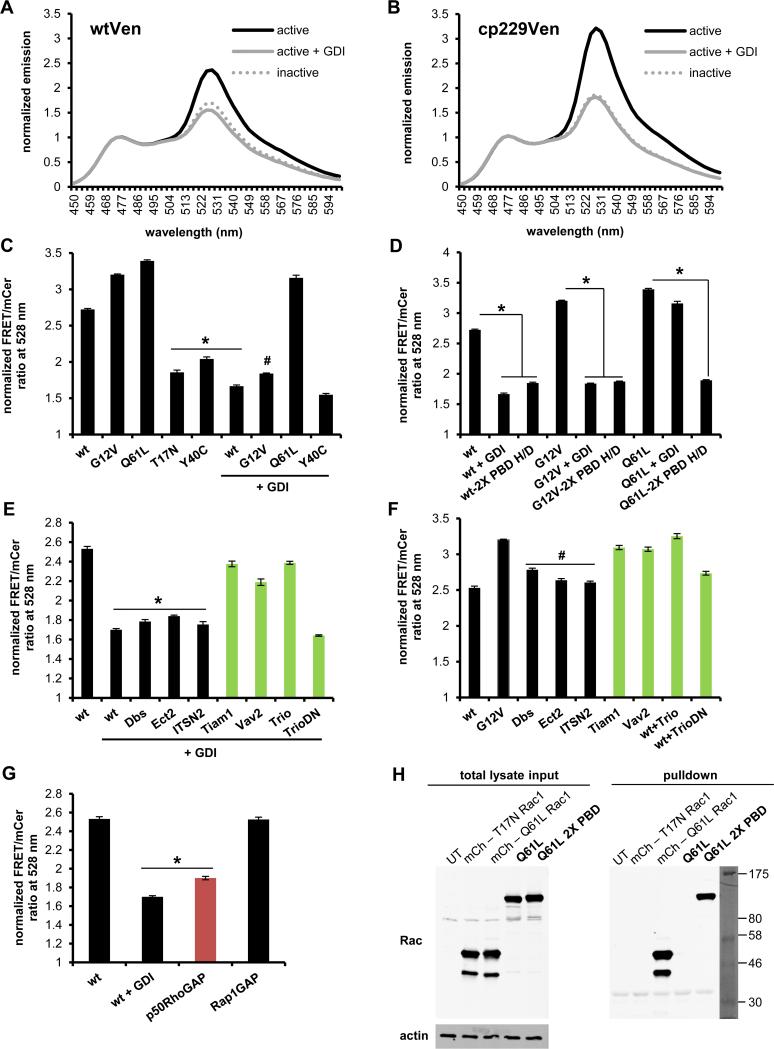
FIGURE 2.
Characterization of new Rac2 biosensor. Normalized emission spectra of constitutively active (G12V) with or without excess GDI and dominant negative (T17N) Rac2 sensor containing A) original mVen B) mcp229Ven, normalized to mCer1 donor emission peak at 474 nm. C) WT or mutant GTPase versions of mcp229Ven Rac2 biosensor in the presence or absence of excess GDI. D) WT and mutant versions of Rac2 biosensor with or without the GTPase-binding deficient H83D, H86D mutations in PBD1 (2X PBD H/D = both PBD1 and PBD2 are GTPase-binding deficient) in the presence or absence of excess GDI. E) WT GTPase version of mcp229Ven biosensor co-expressed with Rac2-targeting (in green) or non-targeting GEFs in the presence or F) absence of excess GDI. G) WT GTPase version of mcp229Ven biosensor co-expressed with excess of targeting (in red) or non-targeting GAPs. H) Western blot of GST-PAK pulldown of mCherry-tagged Q16L and T17N Rac1 for assay control, and Q61L constitutively active version of the Rac2 biosensor with functional or GTPase-binding-deficient mutations in the PBD1 domain, overexpressed in HEK293 cells. Total lysates and actin was used as a loading control. Fluorometry data, performed in HEK293 cells, are the mean +/− SEM of 3 independent experiments performed in triplicates. * p < 0.00001 vs WT alone; # p < 0.00001 vs G12V alone.