Figure 7. Revised model of the translocation mechanism depicting early and late events in the process.
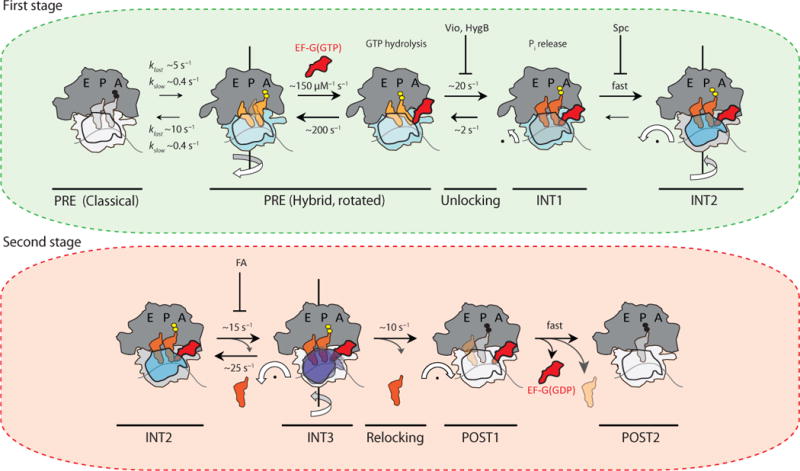
(a) In the first stage of translocation, EF-G(GTP) engages rotated (light blue) configurations of the dynamic PRE complex to stabilize canonical tRNA hybrid (P/E; A/P) positions (light orange). Unrotated (light grey) PRE complexes must spontaneously rotate prior to translocation. GTP hydrolysis and unlocking lead to an intermediate displaying compacted tRNA positions (dark orange) prior to head domain swivel (INT1). These steps are accompanied by inorganic phosphate (Pi) release. INT1 rapidly converts into the relatively stable intermediate (INT2) displaying pronounced (18–21°) head swivel (dark blue) and partial reverse rotation of the small subunit body domain (grey). The inhibitory impacts of viomycin (Vio), hygromycin B (HygB) and spectinomycin (Spc) are indicated in this context, while having little or no effect on Pi release37,48. (b) In the second stage of translocation, exaggerated head domain motions (purple) lead to rapid, reversible excursions of INT2 to a third transient intermediate (INT3). Deacylated tRNA may be released during this exchange. From INT3, the head domain relocks to achieve its unswiveled position (white). In the process mRNA and peptidyl-tRNA are rectified to their POST configurations, followed immediately by EF-G(GDP) release from the ribosome. The rates depicted in this model represent estimates based on the two distinct PRE complexes examined. The values shown for EF-G binding to, and non-productive release from, the PRE complex were taken from ref. 12 and references therein.
