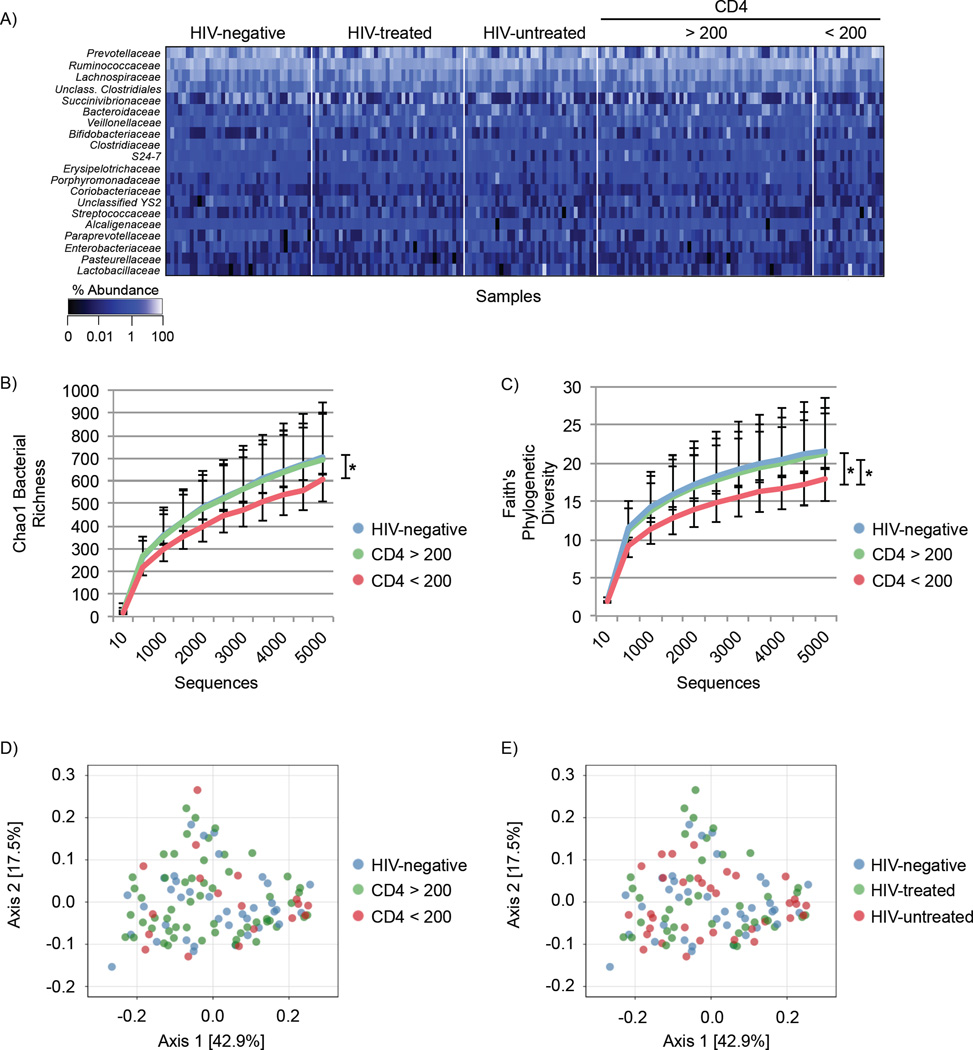
Figure 4. Bacterial community profiling.
(A) Heatmap showing relative abundance of the 20 most frequent bacterial families (y-axis) by sample (x-axis), grouped by HIV status and CD4 T cell count. Percent abundance is indicated by the gradient key. (B) Chao1 rarefied bacterial richness grouped by CD4 T cell count. (C) Comparison of bacterial Faith’s phylogenetic diversity in HIV-negative and HIV-positive subjects by CD4 T cell count. Statistical analysis was performed in QIIME using two-sample, non-parametric t-tests with Monte Carlo permutations. Error bars indicate SEM. p ≤ 0.05 = * Principle Coordinate Analysis (PCoA) plots of the weighted UniFrac distances colored by HIV status and (D) CD4 T cell count or (E) ART treatment. See also Figure S3, S4, Table S7.