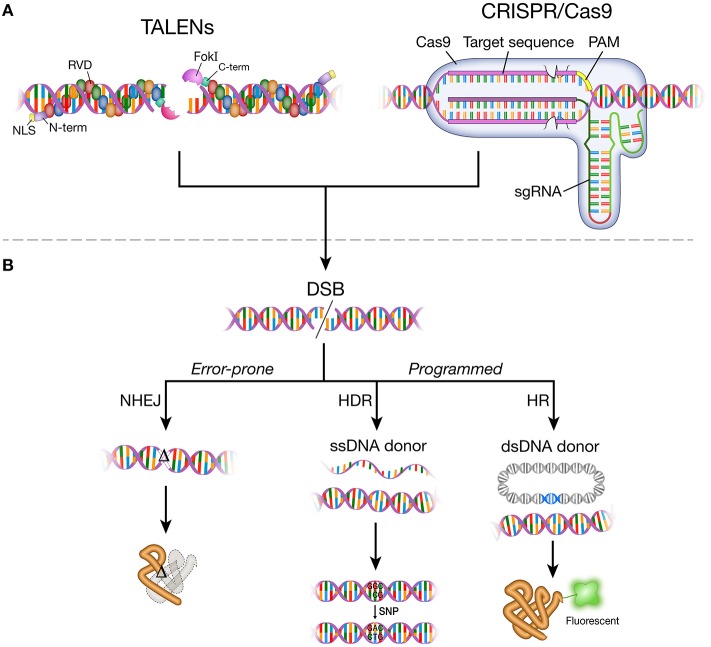
Figure 1.
DNA-binding agents, DNA double-strand break (DSB) and repair pathways. (A) A pair of TALEN monomers bound to double-strand DNA. A TALEN monomer comprises N-terminal domain, nuclear localization signal, modular repeats that contain two highly variable amino acid residues (RVDs), C-terminal domain, and FokI endonuclease. TALENs bind to target DNA in the major groove. A pair of TALEN monomers targeted closely enable FokI dimerization to make DNA double-strand break. Cas9 protein and sgRNA form a complex. The sgRNA-Cas9 complex bind to target DNA through Watson-Crick base-pairing between the target recognition region of the sgRNA and the target DNA sequence (protospacer). The target recognition region (pink rectangle) is the RNA sequence in the sgRNA that matches the target DNA sequence (20 nt). The PAM (protospacer adjacent motif) sequence varies depending on the type of Cas protein. Streptococcus pyogenes Cas9 protein recognizes 5′-NGG-3′ sequence. (B) Targetable TALEN and CRISPR/Cas9 systems generate DNA double-strand break (DSB) at the target locus in the genome. DSBs are repaired by non-homologous end joining (NHEJ), homology-directed repair (HDR), or homologous recombination (HR). Error-prone NHEJ results in non-functional proteins or loss of protein through frameshift mutations introduced to open reading frame. HDR and HR mediate faithful repair of DNA break by incorporating exogenously provided DNA templates in either single strand DNA oligos (ssDNA donor) or plasmids (dsDNA donor). HDR and HR can be used to change small sequences or tag the target gene with a reporter (e.g., green fluorescent protein). TALENs, transcription activator-like effector nucleases; NLS, nuclear localization signal; RVD, repeat variable di-residue; FokI, FokI endonuclease; N-term, N-terminal domain; C-term, C-terminal domain; CRISPR/Cas9, clustered regularly interspaced short palindromic repeats/CRISPR associated protein 9; sgRNA, single guide RNA; PAM, protospacer adjacent motif.