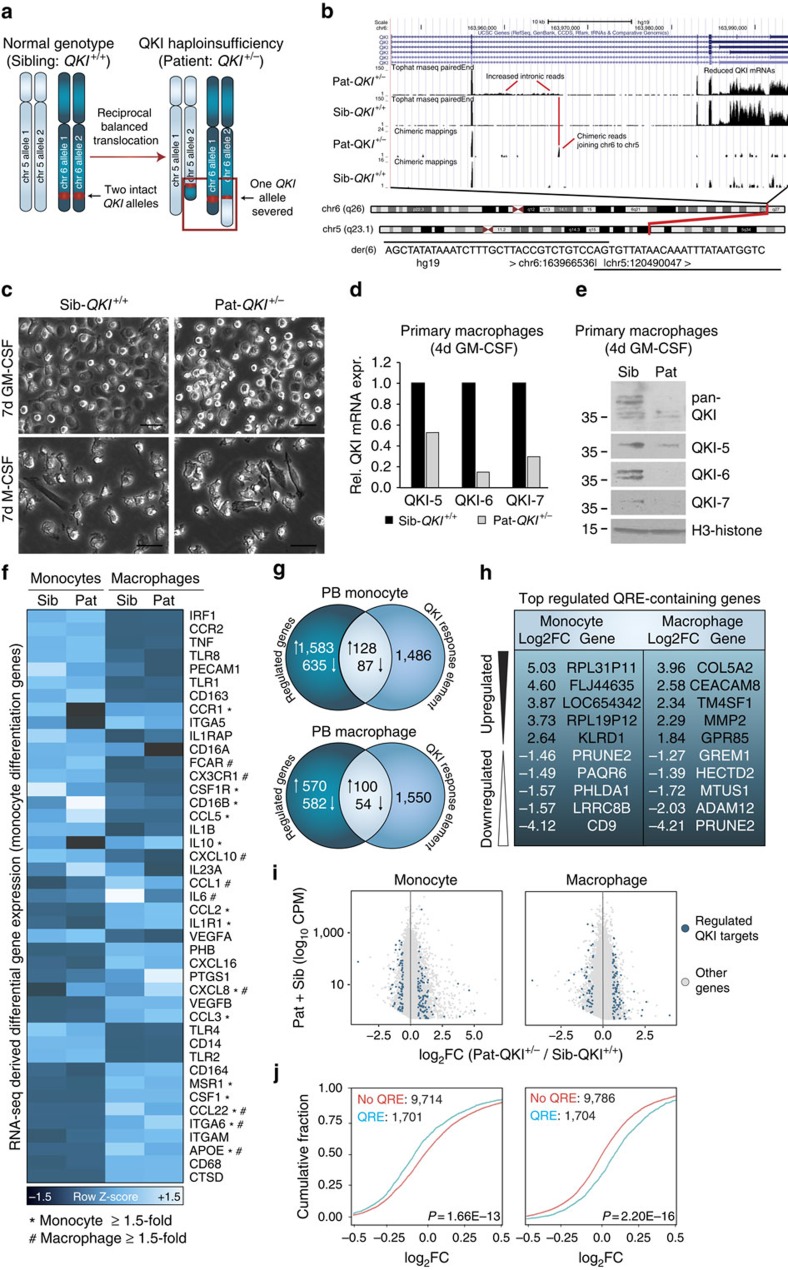
Figure 3. Characterization of monocyte and macrophage biology in a unique QKI haploinsufficient patient.
(a) Schematic of chromosomal translocation event in the qkI haploinsufficient patient (Pat-QKI+/−), reducing QKI expression to ∼50% that of her sister control (Sib-QKI+/+). (b) Top: UCSC Genome Browser display of reference genome QKI locus with standard and chimeric reads for the patient and sibling. The reduced expression levels and altered 3′-untranslated region (UTR) composition in the patient RNA as compared with a sibling control is noteworthy. Patient shows increased intronic RNA extending to the point where chimeric reads map at the breakpoint to chr5. Middle: chromosome diagrams showing normal chromosomes 5 and 6 with the red line, indicating the location of the breakage fusion point. Bottom: sequence across the fusion point. The chromosomal origin of the AG dinucleotide is ambiguous. (c) Photomicrographs of sibling and patient macrophages, cultured in GM-CSF or M-CSF for 7 days, respectively. (d,e) Assessment of QKI isoform mRNA and protein expression in 4-day GM-CSF-stimulated macrophages in the sibling and patient. (f) Hierarchical clustering (Euclidean algorithm) of key monocyte differentiation genes depicting changes in RNA-seq-derived mRNA abundance where dark blue=low expression, whereas light blue=high expression (* and/or # indicates ≥1.5-fold expression change in monocytes or macrophages, respectively). (g) Venn diagrams with numbers of differentially expressed genes (minimally ±1.5-fold; patient/sibling expression) for unstimulated (top) and GM-CSF stimulated macrophages (bottom). An expression cutoff (Pat+Sib expression≥1CPM) was applied. (h) The most differentially expressed genes, harbouring a QRE are depicted. (i) Genome-wide scatterplot of mRNA abundance (y axis: Log10 CPM) versus the log2FC (x axis: Patient/sibling CPM) after an expression cutoff (Pat+Sib expression ≥1 CPM) in monocytes (left) and GM-CSF-stimulated macrophages (right). Blue dots indicate QRE-containing transcripts minimally ±1.5-fold differentially expressed. Grey dots do not fulfill these criteria. (j) CDF (y axis) for QKI target (QRE containing: blue line) and non-target (non-QRE containing: cyan line) mRNAs (x axis: log2FC) in monocytes (left) and macrophages (right). Left shift indicates lower expression of QKI target genes, whereas a right shift indicates higher expression of QKI targets in the patient samples. Distributions were compared using a Wilcoxon rank-sum test.