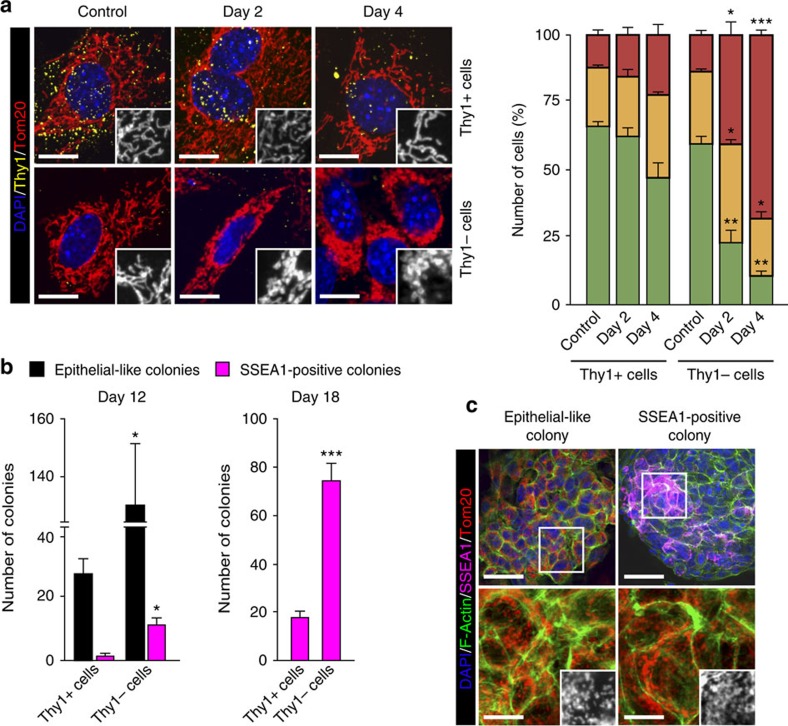
Figure 2. Mitochondrial fission is associated with cells undergoing reprogramming.
(a) MEFs were mock-infected (control) or transduced with the reprogramming factors. At the indicated days, cells were fixed and Thy1 expression and mitochondrial morphology assessed by IF. (left) Representative confocal images of MEFs stained with anti-Thy1 (yellow) and anti-Tom20 (red) antibodies before (control) or after expressing the OSKM factors for the indicated days. Insets show a black and white magnification of the pictures. DAPI (blue) was used as a nuclear counterstaining. Scale bar, 16 μm. Graph on the right shows the quantification of the different mitochondrial morphologies observed in MEFs before (control) or at the indicated days after OSKM expression in the Thy1-positive and -negative cells (n=3). (b) MEFs were mock-infected (control) or transduced with the reprogramming factors. At day 4, cells were sorted as Thy1-positive and -negative. Sorted Thy1-positive and -negative cell populations were further cultured for 8 or 14 days, and epithelial-like and SSEA1-positive colonies were identified by IF analysis using Alexa Fluor 488-Phalloidin and anti-SSEA1 antibody. Graph shows the quantification of epithelial-like and SSEA1-positive colonies found at the indicated days of reprogramming (sorted at day 4) (n=3). (c) Representative confocal images of epithelial-like and SSEA1-positive colonies found in OSKM-transduced MEFs at days 12 and 18 of reprogramming stained with Alexa Fluor 488-Phalloidin (green, to stain F-actin), anti-Tom20 (red, to label mitochondria) and anti-SSEA1 (violet, to label pluripotent colonies) antibodies. (lower) Higher magnifications of the indicated area in the respective upper images. Insets in the lower images are a black and white magnification of the pictures to illustrate mitochondrial morphology in colonies. Scale bar, 24 μm (upper); scale bar, 8 μm (lower), respectively. Data are represented as mean±s.e.m. (*P<0.05, **P<0.01, ***P<0.001). One-tailed unpaired Student's t-test was used to compare data sets.