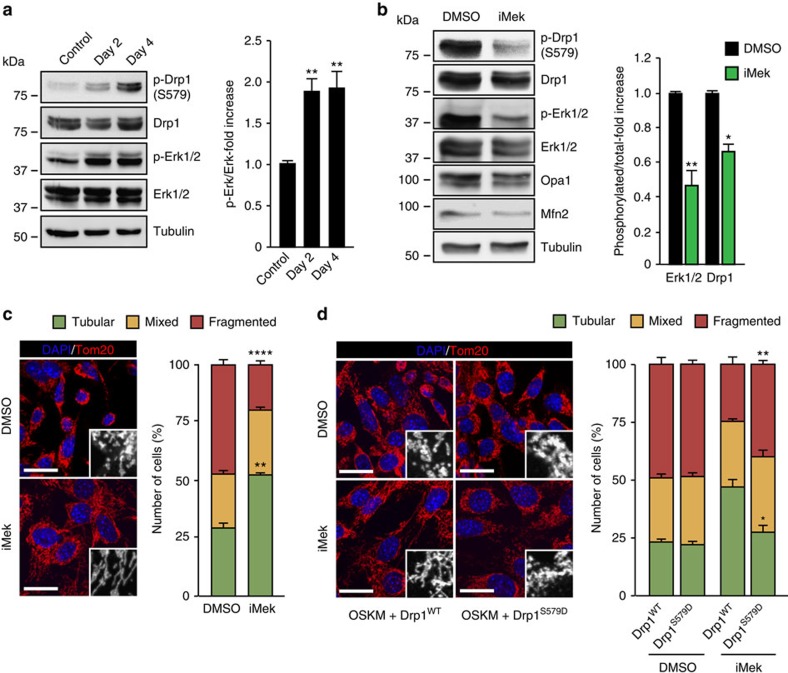
Figure 5. Reprogramming-induced mitochondrial fission depends on Erk1/2 phosphorylation of Drp1.
(a) Lysates of mock- or OSKM-transduced MEFs for the indicated days were analysed by immunoblotting using the indicated antibodies. Graphs on the right show the quantification of the indicated ratios (n=3). (b) MEFs were OSKM-transduced and 3 days post-infection cells were treated with DMSO (black bars), as vehicle control, or the MEK1/2 inhibitor PD0325901 (1 μM) (iMek, green bars) for 16 h. Then, cell lysates were prepared and analysed by immunoblotting using the indicated antibodies (left). Graphs on the right show the quantification of the indicated ratios (n=3). (c) (left) Representative confocal images of OSKM-expressing MEFs for 3 days, treated as in b and stained with anti-Tom20 antibody (red) to assess the different mitochondrial morphologies. Insets show a black and white magnification of the pictures. DAPI (blue) was used as a nuclear counterstaining. Scale bars, 24 μm. Graph on the right shows the quantification of the indicated mitochondrial morphologies observed in the cells treated as indicated (n=3). (d) (left) Representative confocal images of MEFs expressing the reprogramming factors, together with Drp1 wild type (Drp1WT) or the phosphomimetic S579D mutation (Drp1S579D), during 4 days. Cells were then treated, fixed and stained as in c. Insets show a black and white magnification of the pictures. DAPI (blue) was used as a nuclear counterstaining. Scale bars, 24 μm. Graph on the right shows the quantification of the indicated mitochondrial morphologies observed in the cells treated as indicated (n=3). Data are represented as mean±s.e.m. (*P<0.05, **P<0.01, ****P<0.0001). One-tailed unpaired Student's t-test was used to compare data sets.