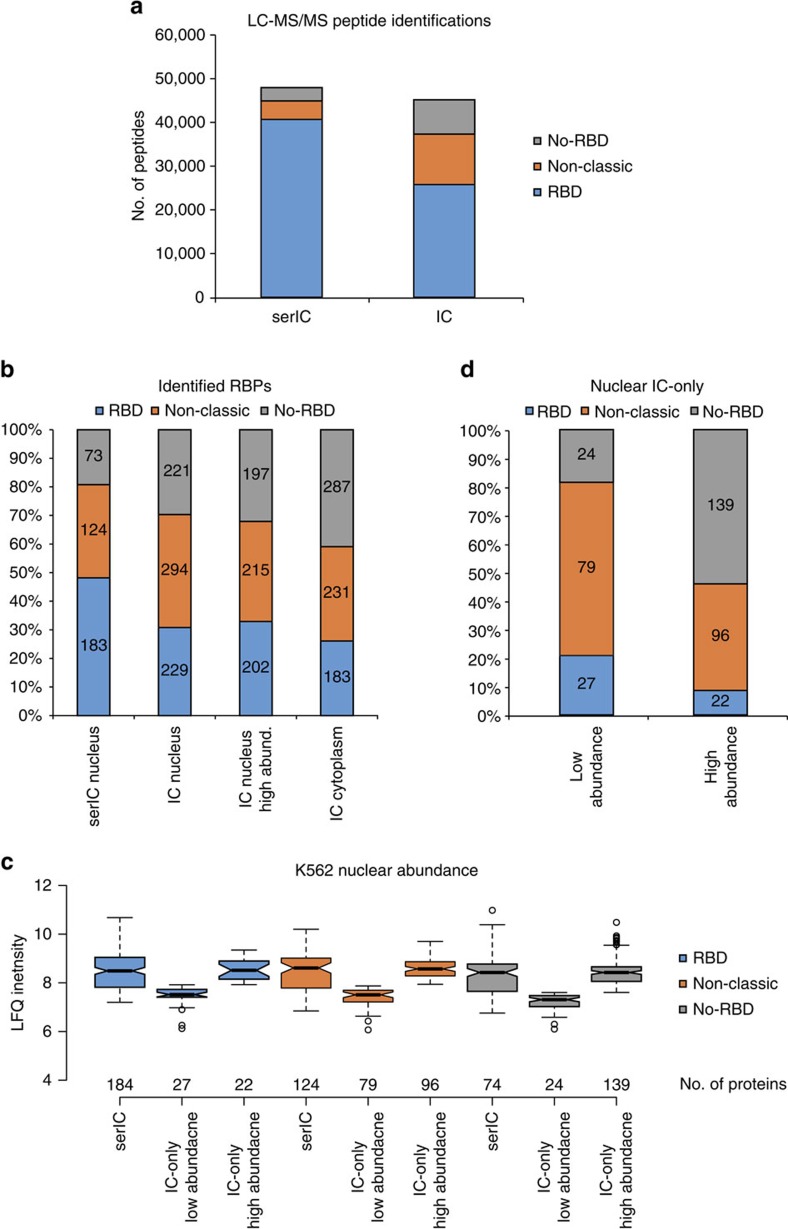
Figure 2. Altered domain composition of the serIC RNA interactome.
(a) The combined number of individual LC–MS/MS peptide identifications in all serIC replicates and in all IC replicates from HeLa15. Indicated are peptides from proteins with classic, non-classic or no RBD. Only peptides from proteins that are enriched over the control are shown for each experiment. (b) Domain composition of the nuclear RNA interactome. Proteins identified by IC in HeLa15 and HEK293 (ref. 14) were filtered for expression in K562 nuclei or cytoplasm. All nuclear, only highly abundant nuclear, and cytoplasmic IC-derived candidates are included in the comparison, respectively. (c) K562 nuclear abundance of the RBPs detected by serIC or in IC only, as measured by LC–MS/MS14,15. IC-only candidates are subdivided into proteins with similar abundance as serIC-derived RBPs from the same class, and proteins with reduced abundance. (d) Distribution of lowly or highly abundant candidate RBPs. Highly abundant IC-only candidates mostly lack RNA-binding domains.